6VWF
 
 | | Structure of ALDH9A1 complexed with NAD+ in space group C222 | | Descriptor: | 4-trimethylaminobutyraldehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Wyatt, J.W, Tanner, J.J. | | Deposit date: | 2020-02-19 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Inhibition, crystal structures, and in-solution oligomeric structure of aldehyde dehydrogenase 9A1.
Arch.Biochem.Biophys., 691, 2020
|
|
7STT
 
 | | Crystal structure of sulfatase from Pedobacter yulinensis | | Descriptor: | CALCIUM ION, CHLORIDE ION, MALONATE ION, ... | | Authors: | O'Malley, A, Schlachter, C.R, Grimes, L.L, Tomashek, J.J, Lee, A.L, Chruszcz, M. | | Deposit date: | 2021-11-15 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.603 Å) | | Cite: | Purification, Characterization, and Structural Studies of a Sulfatase from Pedobacter yulinensis .
Molecules, 27, 2021
|
|
7STU
 
 | | Crystal structure of sulfatase from Pedobacter yulinensis | | Descriptor: | BROMIDE ION, CALCIUM ION, N-acetylgalactosamine-6-sulfatase, ... | | Authors: | O'Malley, A, Schlachter, C.R, Grimes, L.L, Tomashek, J.J, Lee, A.L, Chruszcz, M. | | Deposit date: | 2021-11-15 | | Release date: | 2022-01-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Purification, Characterization, and Structural Studies of a Sulfatase from Pedobacter yulinensis .
Molecules, 27, 2021
|
|
7STV
 
 | | Crystal structure of sulfatase from Pedobacter yulinensis | | Descriptor: | CALCIUM ION, CHLORIDE ION, CITRIC ACID, ... | | Authors: | O'Malley, A, Schlachter, C.R, Grimes, L.L, Tomashek, J.J, Lee, A.L, Chruszcz, M. | | Deposit date: | 2021-11-15 | | Release date: | 2022-01-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Purification, Characterization, and Structural Studies of a Sulfatase from Pedobacter yulinensis .
Molecules, 27, 2021
|
|
2VID
 
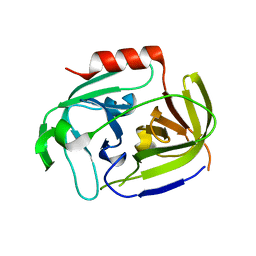 | | Serine protease SplB from Staphylococcus aureus at 1.8A resolution | | Descriptor: | SERINE PROTEASE SPLB | | Authors: | Dubin, G, Stec-Niemczyk, J, Kisielewska, M, Pustelny, K, Popowicz, G.M, Bista, M, Kantyka, T, Boulware, K.T, Stennicke, H.R, Czarna, A, Phopaisarn, M, Daugherty, P.S, Thogersen, I.B, Enghild, J.J, Thornberry, N, Dubin, A, Potempa, J. | | Deposit date: | 2007-11-30 | | Release date: | 2008-05-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Enzymatic Activity of the Staphylococcus Aureus Splb Serine Protease is Induced by Substrates Containing the Sequence Trp-Glu-Leu-Gln.
J.Mol.Biol., 379, 2008
|
|
8FAX
 
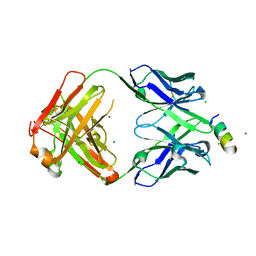 | | Fab 1249A8-MERS Stem Helix Complex | | Descriptor: | 1249A8-HC, 1249A8-LC, CHLORIDE ION, ... | | Authors: | Deshpande, A, Schormann, N, Piepenbrink, M.S, Martinez-Sobrido, L, Kobie, J.J, Walter, M.R. | | Deposit date: | 2022-11-28 | | Release date: | 2023-05-03 | | Last modified: | 2023-07-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and epitope of a neutralizing monoclonal antibody that targets the stem helix of beta coronaviruses.
Febs J., 290, 2023
|
|
2WFD
 
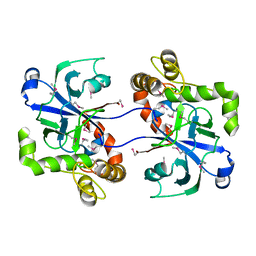 | | Structure of the human cytosolic leucyl-tRNA synthetase editing domain | | Descriptor: | LEUCYL-TRNA SYNTHETASE, CYTOPLASMIC | | Authors: | Seiradake, E, Mao, W, Hernandez, V, Baker, S.J, Plattner, J.J, Alley, M.R.K, Cusack, S. | | Deposit date: | 2009-04-03 | | Release date: | 2009-05-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Crystal Structures of the Human and Fungal Cytosolic Leucyl-tRNA Synthetase Editing Domains: A Structural Basis for the Rational Design of Antifungal Benzoxaboroles.
J.Mol.Biol., 390, 2009
|
|
8FNR
 
 | |
8FNS
 
 | |
1RX0
 
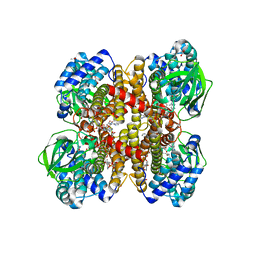 | | Crystal structure of isobutyryl-CoA dehydrogenase complexed with substrate/ligand. | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Acyl-CoA dehydrogenase family member 8, ... | | Authors: | Battaile, K.P, Nguyen, T.V, Vockley, J, Kim, J.J. | | Deposit date: | 2003-12-18 | | Release date: | 2004-04-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structures of Isobutyryl-CoA Dehydrogenase and Enzyme-Product Complex: COMPARISON WITH ISOVALERYL- AND SHORT-CHAIN ACYL-COA DEHYDROGENASES.
J.Biol.Chem., 279, 2004
|
|
8WWX
 
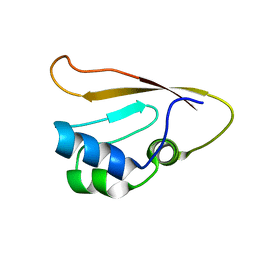 | |
8F88
 
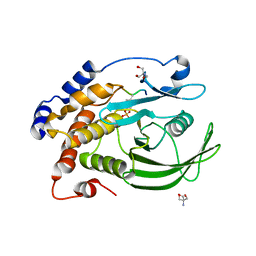 | | Crystal structure of PTP1B D181A/Q262A/C215A phosphatase domain with monophosphorylated JAK2 activation loop phosphopeptide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein kinase JAK2, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Morris, R, Kershaw, N.J, Babon, J.J. | | Deposit date: | 2022-11-21 | | Release date: | 2023-07-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure guided studies of the interaction between PTP1B and JAK.
Commun Biol, 6, 2023
|
|
1S3P
 
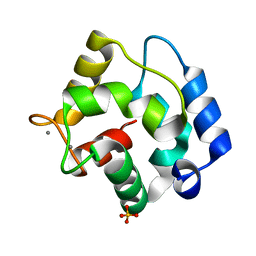 | |
6WB6
 
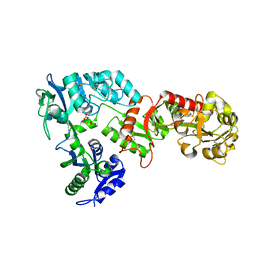 | | 2.05 A resolution structure of transferrin 1 from Manduca sexta | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, FE (III) ION, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Weber, J.J, Gorman, M.J. | | Deposit date: | 2020-03-26 | | Release date: | 2020-11-25 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural insight into the novel iron-coordination and domain interactions of transferrin-1 from a model insect, Manduca sexta.
Protein Sci., 30, 2021
|
|
1BRB
 
 | |
6WAM
 
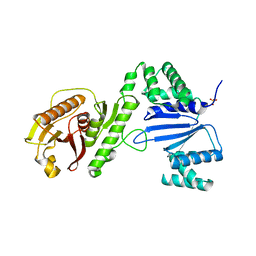 | | Structure of Acinetobacter baumannii Cap4 SAVED/CARF-domain containing receptor | | Descriptor: | SAVED domain-containing protein, SULFATE ION | | Authors: | Lowey, B, Whiteley, A.T, Keszei, A.F.A, Morehouse, B.R, Antine, S.P, Cabrera, V, Schwede, F, Mekalanos, J.J, Shao, S, Lee, A.S.Y, Kranzusch, P.J. | | Deposit date: | 2020-03-25 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | CBASS Immunity Uses CARF-Related Effectors to Sense 3'-5'- and 2'-5'-Linked Cyclic Oligonucleotide Signals and Protect Bacteria from Phage Infection.
Cell, 182, 2020
|
|
1S6W
 
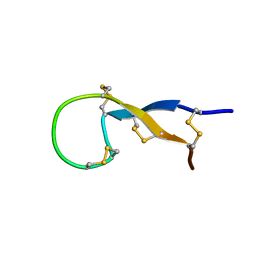 | | Solution Structure of hybrid white striped bass hepcidin | | Descriptor: | Hepcidin | | Authors: | Babon, J.J, Singh, S, Pennington, M.W, Norton, R.S, Westerman, M.E. | | Deposit date: | 2004-01-28 | | Release date: | 2004-12-14 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Bass hepcidin synthesis, solution structure, antimicrobial activities and synergism, and in vivo hepatic response to bacterial infections.
J.Biol.Chem., 280, 2005
|
|
4LMY
 
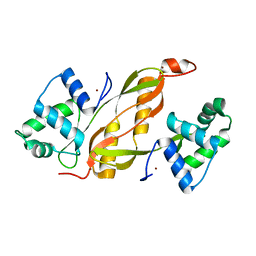 | | Structure of GAS PerR-Zn-Zn | | Descriptor: | Peroxide stress regulator PerR, FUR family, ZINC ION | | Authors: | Lin, C.S, Chao, S.Y, Nix, J.C, Tseng, H.L, Tsou, C.C, Fei, C.H, Ciou, H.S, Jeng, U.S, Lin, Y.S, Chuang, W.J, Wu, J.J, Wang, S. | | Deposit date: | 2013-07-11 | | Release date: | 2014-04-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Distinct structural features of the peroxide response regulator from group a streptococcus drive DNA binding
Plos One, 9, 2014
|
|
6W16
 
 | |
1S98
 
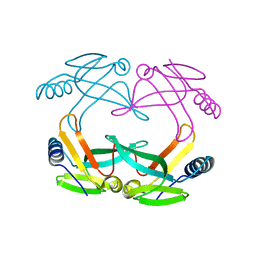 | | E.coli IscA crystal structure to 2.3 A | | Descriptor: | Protein yfhF | | Authors: | Cupp-Vickery, J.R, Silberg, J.J, Ta, D.T, Vickery, L.E. | | Deposit date: | 2004-02-03 | | Release date: | 2004-06-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of IscA, an iron-sulfur cluster assembly protein from Escherichia coli.
J.Mol.Biol., 338, 2004
|
|
2W05
 
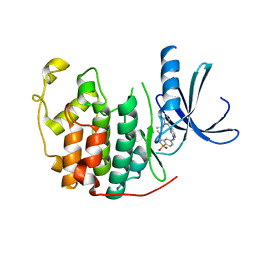 | | Structure of CDK2 in complex with an imidazolyl pyrimidine, compound 5b | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, N-(2-METHOXYETHYL)-4-({4-[2-METHYL-1-(1-METHYLETHYL)-1H-IMIDAZOL-5-YL]PYRIMIDIN-2-YL}AMINO)BENZENESULFONAMIDE | | Authors: | Anderson, M, Andrews, D.M, Barker, A.J, Brassington, C.A, Breed, J, Byth, K.F, Culshaw, J.D, Finlay, M.R, Fisher, E, Green, C.P, Heaton, D.W, Nash, I.A, Newcombe, N.J, Oakes, S.E, Pauptit, R.A, Roberts, A, Stanway, J.J, Thomas, A.P, Tucker, J.A, Weir, H.M. | | Deposit date: | 2008-08-08 | | Release date: | 2008-10-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Imidazoles: Sar and Development of a Potent Class of Cyclin-Dependent Kinase Inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
1RWY
 
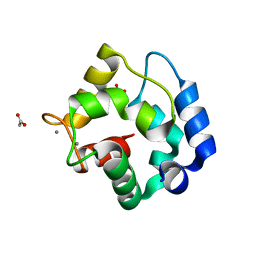 | | CRYSTAL STRUCTURE OF RAT ALPHA-PARVALBUMIN AT 1.05 RESOLUTION | | Descriptor: | ACETIC ACID, AMMONIUM ION, CALCIUM ION, ... | | Authors: | Bottoms, C.A, Schuermann, J.P, Agah, S, Henzl, M.T, Tanner, J.J. | | Deposit date: | 2003-12-17 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Crystal Structure of Rat Alpha-Parvalbumin at 1.05 Resolution
Protein Sci., 13, 2004
|
|
8FOX
 
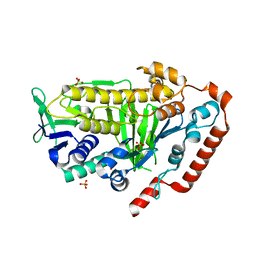 | | AbeH (Tryptophan-5-halogenase) | | Descriptor: | SULFATE ION, Tryptophan 5-halogenase | | Authors: | Ashaduzzaman, M, Bellizzi, J.J. | | Deposit date: | 2023-01-03 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystallographic and thermodynamic evidence of negative cooperativity of flavin and tryptophan binding in the flavin-dependent halogenases AbeH and BorH.
Biorxiv, 2023
|
|
7SUP
 
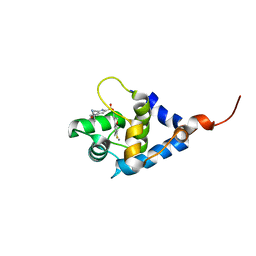 | | NMR structure of cTnC-TnI chimera bound to calcium and A1 | | Descriptor: | 4-(3-cyano-3-methylazetidine-1-carbonyl)-N-[(3S)-7-fluoro-6-methyl-3,4-dihydro-2H-1-benzopyran-3-yl]-5-methyl-1H-pyrrole-2-sulfonamide, CALCIUM ION, Troponin C, ... | | Authors: | Poppe, L, Hartman, J.J, Romero, A, Reagan, J.D. | | Deposit date: | 2021-11-17 | | Release date: | 2022-04-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and Thermodynamic Model for the Activation of Cardiac Troponin.
Biochemistry, 61, 2022
|
|
5M50
 
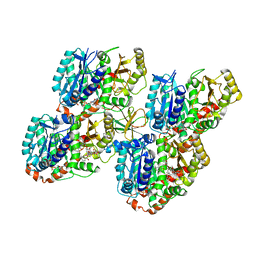 | | Mechanism of microtubule minus-end recognition and protection by CAMSAP proteins | | Descriptor: | Calmodulin-regulated spectrin-associated protein 3, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Akhmanova, A, Moores, C.A, Baldus, M, Steinmetz, M.O, Topf, M, Roberts, A.J, Grant, B.J, Scarabelli, G, Joseph, A.-P, van Hooff, J.J.E, Houben, K, Hua, S, Luo, Y, Stangier, M.M, Jiang, K, Atherton, J. | | Deposit date: | 2016-10-20 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | A structural model for microtubule minus-end recognition and protection by CAMSAP proteins.
Nat. Struct. Mol. Biol., 24, 2017
|
|
