2JDI
 
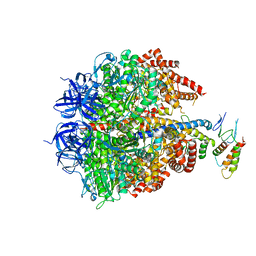 | | Ground state structure of F1-ATPase from bovine heart mitochondria (Bovine F1-ATPase crystallised in the absence of azide) | | Descriptor: | ATP SYNTHASE DELTA CHAIN, ATP SYNTHASE EPSILON CHAIN, ATP SYNTHASE GAMMA CHAIN, ... | | Authors: | Bowler, M.W, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2007-01-09 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ground State Structure of F1-ATPase from Bovine Heart Mitochondria at 1.9 A Resolution
J.Biol.Chem., 282, 2007
|
|
1ZTK
 
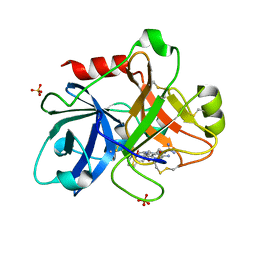 | | Crystal Structure of the Catalytic Domain of Coagulation Factor XI in Complex with 2-(5-Amino-6-oxo-2-m-tolyl-6H-pyrimidin-1-yl)-N-[4-guanidino-1-(thiazole-2-carbonyl)-butyl]-acetamide | | Descriptor: | 2-(5-AMINO-6-OXO-2-M-TOLYL-6H-PYRIMIDIN-1-YL)-N-[4-GUANIDINO-1-(THIAZOLE-2-CARBONYL)-BUTYL]-ACETAMIDE, Coagulation factor XI, SULFATE ION | | Authors: | Nagafuji, P, Jin, L, Rynkiewicz, M, Quinn, J, Bibbins, F, Meyers, H, Babine, R.E, Strickler, J.E, Abdel-Meguid, S.S. | | Deposit date: | 2005-05-27 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Pyrimidinone Inhibitors of a Thrombolytic Protease
To be Published
|
|
289D
 
 | | TARGETING THE MINOR GROOVE OF DNA: CRYSTAL STRUCTURES OF TWO COMPLEXES BETWEEN FURAN DERIVATIVES OF BERENIL AND THE DNA DODECAMER D(CGCGAATTCGCG)2 | | Descriptor: | 2,5-BIS{[4-(N-CYCLOPROPYLDIAMINOMETHYL)PHENYL]}FURAN, DNA (5'-R(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Trent, J.O, Clark, G.R, Kumar, A, Wilson, W.D, Boykin, D.W, Hall, J.E, Tidwell, R.R, Blagburn, B.L, Neidle, S. | | Deposit date: | 1996-10-10 | | Release date: | 1996-12-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Targeting the minor groove of DNA: crystal structures of two complexes between furan derivatives of berenil and the DNA dodecamer d(CGCGAATTCGCG)2.
J.Med.Chem., 39, 1996
|
|
2GRH
 
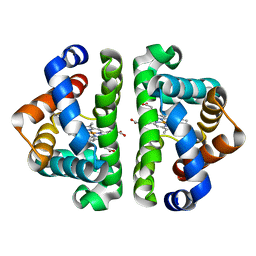 | | M37V mutant of Scapharca dimeric hemoglobin, with CO bound | | Descriptor: | CARBON MONOXIDE, Globin-1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Knapp, J.E, Pahl, R, Srajer, V, Royer Jr, W.E. | | Deposit date: | 2006-04-24 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Allosteric action in real time: Time-resolved crystallographic studies of a cooperative dimeric hemoglobin.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2JNC
 
 | | Refined 3D NMR structure of ECD1 of mCRF-R2beta at pH 5 | | Descriptor: | Corticotropin-releasing factor receptor 2 | | Authors: | Grace, C.R.R, Perrin, M.H, Jozsef, G, DiGruccio, M.R, Cantle, J.P, Rivier, J.E, Vale, W.W, Riek, R. | | Deposit date: | 2007-01-08 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Structure of the N-terminal domain of a type B1 G protein-coupled receptor in complex with a peptide ligand
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2JND
 
 | | 3D NMR structure of ECD1 of mCRF-R2b in complex with Astressin | | Descriptor: | ASTRESSIN, Corticotropin-releasing factor receptor 2 | | Authors: | Grace, C.R.R, Perrin, M.H, Jozsef, G, DiGruccio, M.R, Cantle, J.P, Rivier, J.E, Vale, W.W, Riek, R. | | Deposit date: | 2007-01-08 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Structure of the N-terminal domain of a type B1 G protein-coupled receptor in complex with a peptide ligand
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2JMX
 
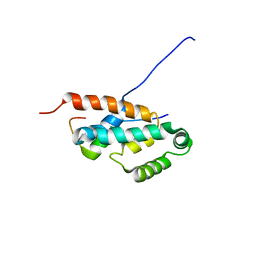 | | OSCP-NT (1-120) in complex with N-terminal (1-25) alpha subunit from F1-ATPase | | Descriptor: | ATP synthase O subunit, mitochondrial, ATP synthase subunit alpha heart isoform | | Authors: | Carbajo, R.J, Neuhaus, D, Kellas, F.A, Yang, J, Runswick, M.J, Montgomery, M.G, Walker, J.E. | | Deposit date: | 2006-12-12 | | Release date: | 2007-04-24 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | How the N-terminal Domain of the OSCP Subunit of Bovine F(1)F(o)-ATP Synthase Interacts with the N-terminal Region of an Alpha Subunit
J.Mol.Biol., 368, 2007
|
|
3ZR5
 
 | | STRUCTURE OF GALACTOCEREBROSIDASE FROM MOUSE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Deane, J.E, Graham, S.C, Kim, N.N, Stein, P.E, Mcnair, R, Cachon-Gonzalez, M.B, Cox, T.M, Read, R.J. | | Deposit date: | 2011-06-14 | | Release date: | 2011-09-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights Into Krabbe Disease from Structures of Galactocerebrosidase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3ZFM
 
 | | Crystal structure of EphB2 | | Descriptor: | EPHRIN TYPE-B RECEPTOR 2 | | Authors: | Debreczeni, J.E, Overman, R, Truman, C, McAlister, M, Attwood, T.K. | | Deposit date: | 2012-12-12 | | Release date: | 2014-01-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Completing the Structural Family Portrait of the Human Ephb Tyrosine Kinase Domains
Protein Sci., 23, 2014
|
|
3ZEW
 
 | | Crystal structure of EphB4 in complex with staurosporine | | Descriptor: | EPHRIN TYPE-B RECEPTOR 4, STAUROSPORINE, SULFATE ION | | Authors: | Debreczeni, J.E, Overman, R, Truman, C, McAlister, M, Attwood, T.K. | | Deposit date: | 2012-12-07 | | Release date: | 2013-12-25 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Completing the Structural Family Portrait of the Human Ephb Tyrosine Kinase Domains
Protein Sci., 23, 2014
|
|
3ZTJ
 
 | | Structure of influenza A neutralizing antibody selected from cultures of single human plasma cells in complex with human H3 Influenza haemagglutinin. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FI6V3 ANTIBODY HEAVY CHAIN, ... | | Authors: | Voss, J.E, Vachieri, S.G, Gamblin, S.J, Collins, P.J, Haire, L.F, Skehel, J.J. | | Deposit date: | 2011-07-08 | | Release date: | 2011-08-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | A Neutralizing Antibody Selected from Plasma Cells that Binds to Group 1 and Group 2 Influenza a Hemagglutinins.
Science, 333, 2011
|
|
3ZLC
 
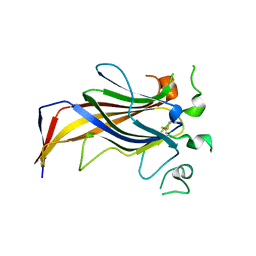 | | Crystal Structure of Erv41p | | Descriptor: | ER-DERIVED VESICLES PROTEIN ERV41 | | Authors: | Biterova, E.I, Svard, M, Possner, D.D.D, Guy, J.E. | | Deposit date: | 2013-01-30 | | Release date: | 2013-03-27 | | Last modified: | 2013-06-12 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | The Crystal Structure of the Lumenal Domain of Erv41P, a Protein Involved in Transport between the Endoplasmic Reticulum and Golgi Apparatus
J.Mol.Biol., 425, 2013
|
|
3ZGP
 
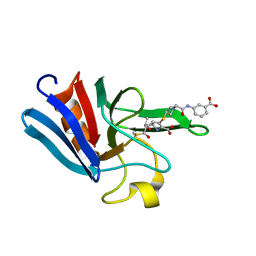 | | NMR structure of the catalytic domain from E. faecium L,D- transpeptidase acylated by ertapenem | | Descriptor: | (4R,5S)-3-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-4-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, ERFK/YBIS/YCFS/YNHG | | Authors: | Lecoq, L, Triboulet, S, Dubee, V, Bougault, C, Hugonnet, J.E, Arthur, M, Simorre, J.P. | | Deposit date: | 2012-12-18 | | Release date: | 2013-04-24 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Structure of Enterococcus Faecium L,D---Transpeptidase Acylated by Ertapenem Provides Insight Into the Inactivation Mechanism.
Acs Chem.Biol., 8, 2013
|
|
3ZR6
 
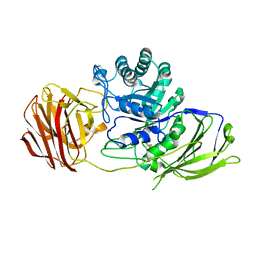 | | STRUCTURE OF GALACTOCEREBROSIDASE FROM MOUSE IN COMPLEX WITH GALACTOSE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Deane, J.E, Graham, S.C, Kim, N.N, Stein, P.E, Mcnair, R, Cachon-Gonzalez, M.B, Cox, T.M, Read, R.J. | | Deposit date: | 2011-06-14 | | Release date: | 2011-09-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Insights Into Krabbe Disease from Structures of Galactocerebrosidase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3ZOY
 
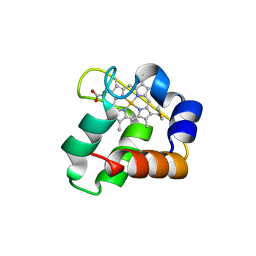 | | Crystal Structure of N64Del Mutant of Nitrosomonas europaea Cytochrome c552 (hexagonal space group) | | Descriptor: | CYTOCHROME C-552, HEME C | | Authors: | Hersleth, H.-P, Can, M, Krucinska, J, Zoppellaro, G, Andersen, N.H, Wedekind, J.E, Andersson, K.K, Bren, K.L. | | Deposit date: | 2013-02-26 | | Release date: | 2013-08-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Characterization of Nitrosomonas Europaea Cytochrome C-552 Variants with Marked Differences in Electronic Structure.
Chembiochem, 14, 2013
|
|
3ZFX
 
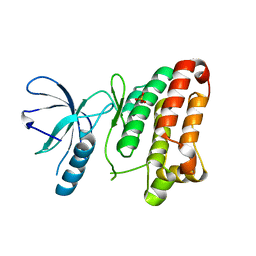 | | Crystal structure of EphB1 | | Descriptor: | EPHRIN TYPE-B RECEPTOR 1, SULFATE ION | | Authors: | Debreczeni, J.E, Overman, R, Truman, C, McAlister, M, Attwood, T.K. | | Deposit date: | 2012-12-12 | | Release date: | 2014-01-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Completing the Structural Family Portrait of the Human Ephb Tyrosine Kinase Domains
Protein Sci., 23, 2014
|
|
1SD1
 
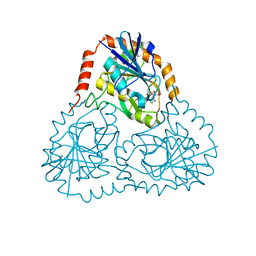 | | STRUCTURE OF HUMAN 5'-DEOXY-5'-METHYLTHIOADENOSINE PHOSPHORYLASE COMPLEXED WITH FORMYCIN A | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, 5'-methylthioadenosine phosphorylase | | Authors: | Lee, J.E, Settembre, E.C, Cornell, K.A, Riscoe, M.K, Sufrin, J.R, Ealick, S.E, Howell, P.L. | | Deposit date: | 2004-02-12 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural Comparison of MTA Phosphorylase and MTA/AdoHcy Nucleosidase Explains Substrate Preferences and Identifies Regions Exploitable for Inhibitor Design.
Biochemistry, 43, 2004
|
|
1SD2
 
 | | STRUCTURE OF HUMAN 5'-DEOXY-5'-METHYLTHIOADENOSINE PHOSPHORYLASE COMPLEXED WITH 5'-METHYLTHIOTUBERCIDIN | | Descriptor: | 2-(4-AMINO-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-METHYLSULFANYLMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, 5'-methylthioadenosine phosphorylase, SULFATE ION | | Authors: | Lee, J.E, Settembre, E.C, Cornell, K.A, Riscoe, M.K, Sufrin, J.R, Ealick, S.E, Howell, P.L. | | Deposit date: | 2004-02-12 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Comparison of MTA Phosphorylase and MTA/AdoHcy Nucleosidase Explains Substrate Preferences and Identifies Regions Exploitable for Inhibitor Design.
Biochemistry, 43, 2004
|
|
2QHR
 
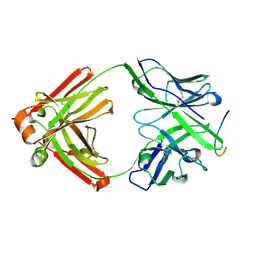 | | Crystal structure of the 13F6-1-2 Fab fragment bound to its Ebola virus glycoprotein peptide epitope. | | Descriptor: | 13F6-1-2 Fab fragment V lambda x light chain, 13F6-1-2 Fab fragment heavy chain, Envelope glycoprotein peptide | | Authors: | Lee, J.E, Kuehne, A, Abelson, D.M, Fusco, M.L, Hart, M.K, Saphire, E.O. | | Deposit date: | 2007-07-02 | | Release date: | 2008-01-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Complex of a protective antibody with its Ebola virus GP peptide epitope: unusual features of a V lambda x light chain.
J.Mol.Biol., 375, 2008
|
|
4EE0
 
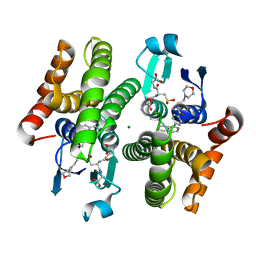 | | Crystal structure of hH-PGDS with water displacing inhibitor | | Descriptor: | 4-(isoquinolin-1-yl)-N-[2-(morpholin-4-yl)ethyl]benzamide, Hematopoietic prostaglandin D synthase, L-GAMMA-GLUTAMYL-3-SULFINO-L-ALANYLGLYCINE, ... | | Authors: | Day, J.E, Thorarensen, A, Trujillo, J.I. | | Deposit date: | 2012-03-28 | | Release date: | 2012-07-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Investigation of the binding pocket of human hematopoietic prostaglandin (PG) D2 synthase (hH-PGDS): a tale of two waters.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4EDT
 
 | | The structure of the S. aureus DnaG RNA Polymerase Domain bound to ppGpp and Manganese | | Descriptor: | BENZAMIDINE, DNA primase, GUANOSINE-5',3'-TETRAPHOSPHATE, ... | | Authors: | Rymer, R.U, Solorio, F.A, Chu, C, Corn, J.E, Wang, J.D, Berger, J.M. | | Deposit date: | 2012-03-27 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Binding Mechanism of Metal-NTP Substrates and Stringent-Response Alarmones to Bacterial DnaG-Type Primases.
Structure, 20, 2012
|
|
4UUD
 
 | | Human dynamin 1 K44A superconstricted polymer stabilized with GTP | | Descriptor: | DYNAMIN-1 | | Authors: | Sundborger, A.C, Fang, S, Heymann, J.A, Ray, P, Chappie, J.S, Hinshaw, J.E. | | Deposit date: | 2014-07-25 | | Release date: | 2014-08-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (12.5 Å) | | Cite: | A Dynamin Mutant Defines a Superconstricted Prefission State.
Cell Rep., 8, 2014
|
|
7OSB
 
 | | Crystal Structure of a Double Mutant PETase (S238F/W159H) from Ideonella sakaiensis | | Descriptor: | CHLORIDE ION, GLYCEROL, Poly(ethylene terephthalate) hydrolase, ... | | Authors: | Shakespeare, T.J, Zahn, M, Allen, M.D, McGeehan, J.E. | | Deposit date: | 2021-06-08 | | Release date: | 2021-10-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Comparative Performance of PETase as a Function of Reaction Conditions, Substrate Properties, and Product Accumulation.
ChemSusChem, 15, 2022
|
|
4EDR
 
 | | The structure of the S. aureus DnaG RNA Polymerase Domain bound to UTP and Manganese | | Descriptor: | BENZAMIDINE, DNA primase, MANGANESE (II) ION, ... | | Authors: | Rymer, R.U, Solorio, F.A, Chu, C, Corn, J.E, Wang, J.D, Berger, J.M. | | Deposit date: | 2012-03-27 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Binding Mechanism of Metal-NTP Substrates and Stringent-Response Alarmones to Bacterial DnaG-Type Primases.
Structure, 20, 2012
|
|
2QJJ
 
 | | Crystal structure of D-Mannonate dehydratase from Novosphingobium aromaticivorans | | Descriptor: | MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme | | Authors: | Fedorov, A.A, Fedorov, E.V, Rakus, J.F, Vick, J.E, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2007-07-07 | | Release date: | 2007-10-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: D-Mannonate dehydratase from Novosphingobium aromaticivorans.
Biochemistry, 46, 2007
|
|
