4B1L
 
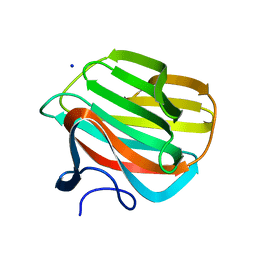 | | CARBOHYDRATE BINDING MODULE CBM66 FROM BACILLUS SUBTILIS | | Descriptor: | LEVANASE, SODIUM ION, beta-D-fructofuranose | | Authors: | Cuskin, F, Flint, J.E, Morland, C, Basle, A, Henrissat, B, Countinho, P.M, Strazzulli, A, Solzehinkin, A, Davies, G.J, Gilbert, H.J, Gloster, T.M. | | Deposit date: | 2012-07-11 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | How Nature Can Exploit Nonspecific Catalytic and Carbohydrate Binding Modules to Create Enzymatic Specificity
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1NF9
 
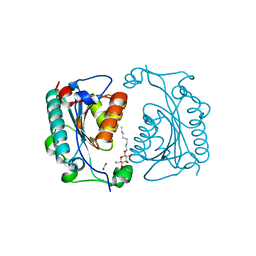 | | Crystal Structure of PhzD protein from Pseudomonas aeruginosa | | Descriptor: | FORMIC ACID, octyl beta-D-glucopyranoside, phenazine biosynthesis protein phzD | | Authors: | Parsons, F, Calabrese, K, Eisenstein, E, Ladner, J.E. | | Deposit date: | 2002-12-13 | | Release date: | 2003-06-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and mechanism of Pseudomonas aeruginosa PhzD, an isochorismatase from the phenazine biosynthetic pathway
Biochemistry, 42, 2003
|
|
1NF8
 
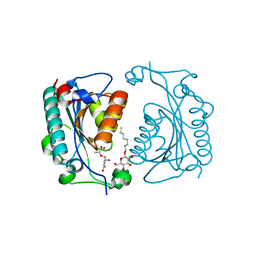 | | Crystal structure of PhzD protein active site mutant with substrate | | Descriptor: | (5S,6S)-5-[(1-carboxyethenyl)oxy]-6-hydroxycyclohexa-1,3-diene-1-carboxylic acid, octyl beta-D-glucopyranoside, phenazine biosynthesis protein phzD | | Authors: | Parsons, F, Calabrese, K, Eisenstein, E, Ladner, J.E. | | Deposit date: | 2002-12-13 | | Release date: | 2003-06-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and mechanism of Pseudomonas aeruginosa PhzD, an isochorismatase from the phenazine biosynthetic pathway
Biochemistry, 42, 2003
|
|
2P58
 
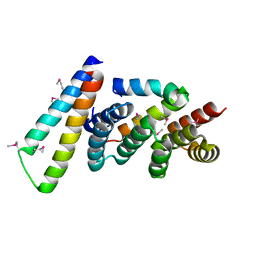 | | Structure of the Yersinia pestis Type III secretion system needle protein YscF in complex with its chaperones YscE/YscG | | Descriptor: | Putative type III secretion protein YscE, Putative type III secretion protein YscF, Putative type III secretion protein YscG | | Authors: | Sun, P, Austin, B.P, Tropea, J.E, Waugh, D.S. | | Deposit date: | 2007-03-14 | | Release date: | 2008-03-04 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural characterization of the Yersinia pestis type III secretion system needle protein YscF in complex with its heterodimeric chaperone YscE/YscG.
J.Mol.Biol., 377, 2008
|
|
3I2B
 
 | | The crystal structure of human 6 Pyruvoyl Tetrahydrobiopterin Synthase | | Descriptor: | 1,2-ETHANEDIOL, 6-pyruvoyl tetrahydrobiopterin synthase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ugochukwu, E, Cocking, R, Pilka, E, Yue, W.W, Bray, J.E, Chaikuad, A, Krojer, T, Muniz, J, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-06-29 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of human 6 Pyruvoyl Tetrahydrobiopterin Synthase
To be Published
|
|
2LK3
 
 | | U2/U6 Helix I | | Descriptor: | RNA (5'-R(*GP*GP*CP*UP*UP*AP*GP*AP*UP*CP*AP*GP*AP*AP*AP*UP*GP*AP*UP*CP*AP*GP*CP*C)-3') | | Authors: | Burke, J.E, Sashital, D.G, Zuo, X.E, Wang, Y, Butcher, S.E. | | Deposit date: | 2011-10-03 | | Release date: | 2012-02-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the yeast U2/U6 snRNA complex.
Rna, 18, 2012
|
|
3JWD
 
 | | Structure of HIV-1 gp120 with gp41-Interactive Region: Layered Architecture and Basis of Conformational Mobility | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB 48D HEAVY CHAIN, FAB 48D LIGHT CHAIN, ... | | Authors: | Pancera, M, Majeed, S, Ban, Y.A, Chen, L, Huang, C.C, Kong, L, Kwon, Y.D, Stuckey, J, Zhou, T, Robinson, J.E, Schief, W.R, Sodroski, J, Wyatt, R, Kwong, P.D. | | Deposit date: | 2009-09-18 | | Release date: | 2009-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2L00
 
 | | Solution structure of the non-covalent complex of the ZNF216 A20 domain with ubiquitin | | Descriptor: | Ubiquitin, ZINC ION, Zfand5 protein (Zinc finger protein 216 (Predicted), ... | | Authors: | Garner, T.P, Long, J.E, Searle, M.S, Layfield, R. | | Deposit date: | 2010-06-29 | | Release date: | 2011-07-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Co-localisation of ubiquitin receptors ZNF216 and p62 in a ubiquitin-mediated ternary complex
To be Published
|
|
4DMX
 
 | | Cathepsin K inhibitor | | Descriptor: | (1R,2R)-N-(1-cyanocyclopropyl)-2-{[4-(4-fluorophenyl)piperazin-1-yl]carbonyl}cyclohexanecarboxamide, Cathepsin K, GLYCEROL | | Authors: | Dossetter, A.G, Beeley, H, Bowyer, J, Cook, C.R, Crawford, J.J, Finlayson, J.E, Heron, N.M, Heyes, C, Highton, A.J, Hudson, J.A, Kenny, P.W, Martin, S, MacFaul, P.A, McGuire, T.M, Gutierrez, P.M, Morley, A.D, Morris, J.J, Page, K.M, Rosenbrier Ribeiro, L, Sawney, H, Steinbacher, S, Krapp, S, Jestel, A, Smith, C, Vickers, M. | | Deposit date: | 2012-02-08 | | Release date: | 2012-07-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | (1R,2R)-N-(1-cyanocyclopropyl)-2-(6-methoxy-1,3,4,5-tetrahydropyrido[4,3-b]indole-2-carbonyl)cyclohexanecarboxamide (AZD4996): a potent and highly selective cathepsin K inhibitor for the treatment of osteoarthritis.
J.Med.Chem., 55, 2012
|
|
4DMY
 
 | | Cathepsin K inhibitor | | Descriptor: | (1R,2R)-N-(1-cyanocyclopropyl)-2-[(8-fluoro-1,3,4,5-tetrahydro-2H-pyrido[4,3-b]indol-2-yl)carbonyl]cyclohexanecarboxamide, Cathepsin K, GLYCEROL, ... | | Authors: | Dossetter, A.G, Beeley, H, Bowyer, J, Cook, C.R, Crawford, J.J, Finlayson, J.E, Heron, N.M, Heyes, C, Highton, A.J, Hudson, J.A, Kenny, P.W, Martin, S, MacFaul, P.A, McGuire, T.M, Gutierrez, P.M, Morley, A.D, Morris, J.J, Page, K.M, Rosenbrier Ribeiro, L, Sawney, H, Steinbacher, S, Krapp, S, Jestel, A, Smith, C, Vickers, M. | | Deposit date: | 2012-02-08 | | Release date: | 2012-07-11 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | (1R,2R)-N-(1-cyanocyclopropyl)-2-(6-methoxy-1,3,4,5-tetrahydropyrido[4,3-b]indole-2-carbonyl)cyclohexanecarboxamide (AZD4996): a potent and highly selective cathepsin K inhibitor for the treatment of osteoarthritis.
J.Med.Chem., 55, 2012
|
|
1ONE
 
 | | YEAST ENOLASE COMPLEXED WITH AN EQUILIBRIUM MIXTURE OF 2'-PHOSPHOGLYCEATE AND PHOSPHOENOLPYRUVATE | | Descriptor: | 2-PHOSPHOGLYCERIC ACID, ENOLASE, MAGNESIUM ION, ... | | Authors: | Larsen, T.M, Wedekind, J.E, Rayment, I, Reed, G.H. | | Deposit date: | 1995-12-05 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A carboxylate oxygen of the substrate bridges the magnesium ions at the active site of enolase: structure of the yeast enzyme complexed with the equilibrium mixture of 2-phosphoglycerate and phosphoenolpyruvate at 1.8 A resolution.
Biochemistry, 35, 1996
|
|
2OUE
 
 | |
2OT5
 
 | |
4DKT
 
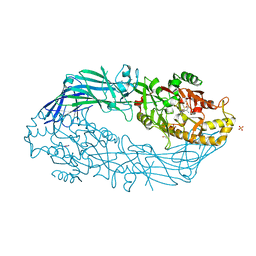 | | Crystal structure of human peptidylarginine deiminase 4 in complex with N-acetyl-L-threonyl-L-alpha-aspartyl-N5-[(1E)-2-fluoroethanimidoyl]-L-ornithinamide | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Protein-arginine deiminase type-4, ... | | Authors: | Jones, J.E, Slack, J.L, Fang, P, Zhang, X, Subramanian, V, Causey, C.P, Coonrod, S.A, Guo, M, Thompson, P.R. | | Deposit date: | 2012-02-04 | | Release date: | 2012-02-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Synthesis and Screening of a Haloacetamidine Containing Library To Identify PAD4 Selective Inhibitors.
Acs Chem.Biol., 7, 2012
|
|
3B5A
 
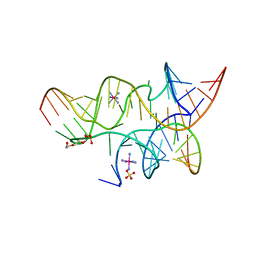 | | Crystal Structure of a Minimally Hinged Hairpin Ribozyme Incorporating A38G mutation with a 2'OMe modification at the active site | | Descriptor: | 29-mer Loop A and Loop B Ribozyme strand, COBALT HEXAMMINE(III), Loop A Substrate strand, ... | | Authors: | MacElrevey, C, Krucinska, J, Wedekind, J.E. | | Deposit date: | 2007-10-25 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural effects of nucleobase variations at key active site residue Ade38 in the hairpin ribozyme.
Rna, 14, 2008
|
|
1JZM
 
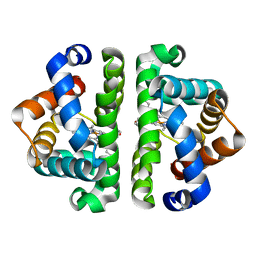 | | Crystal Structure of Scapharca inaequivalvis HbI, I114M Mutant in the Absence of ligand. | | Descriptor: | GLOBIN I - ARK SHELL, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Knapp, J.E, Gibson, Q.H, Cushing, L, Royer Jr, W.E. | | Deposit date: | 2001-09-16 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Restricting the Ligand-Linked Heme Movement in Scapharca Dimeric Hemoglobin Reveals Tight Coupling between Distal and Proximal
Contributions to Cooperativity.
Biochemistry, 40, 2001
|
|
2Y96
 
 | |
1LR1
 
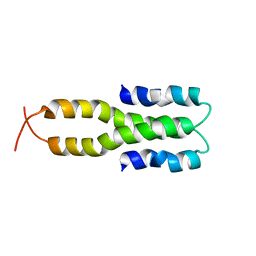 | | Solution Structure of the Oligomerization Domain of the Bacterial Chromatin-Structuring Protein H-NS | | Descriptor: | dna-binding protein h-ns | | Authors: | Esposito, D, Petrovic, A, Harris, R, Ono, S, Eccleston, J, Mbabaali, A, Haq, I, Higgins, C.F, Hinton, J.C.D, Driscoll, P.C, Ladbury, J.E. | | Deposit date: | 2002-05-14 | | Release date: | 2003-01-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | H-NS Oligomerization Domain Structure Reveals the Mechanism for High Order
Self-association of the Intact Protein
J.Mol.Biol., 324, 2002
|
|
1Q3E
 
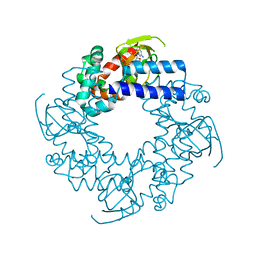 | | HCN2J 443-645 in the presence of cGMP | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 | | Authors: | Zagotta, W.N, Olivier, N.B, Black, K.D, Young, E.C, Olson, R, Gouaux, J.E. | | Deposit date: | 2003-07-29 | | Release date: | 2003-09-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for modulation and agonist specificity of HCN pacemaker channels
Nature, 425, 2003
|
|
2YG0
 
 | | CBM62 FROM CLOSTRIDIUM THERMOCELLUM XYL5A | | Descriptor: | CALCIUM ION, CARBOHYDRATE BINDING FAMILY 6, GLYCEROL, ... | | Authors: | Montanier, C.Y, Correia, M.A.S, Flint, J.E, Zhu, Y, Basle, A, Mckee, L.S, Prates, J.A.M, Polizzi, S.J, Coutinho, P.M, Henrissat, B, Fontes, C.M.G.A, Gilbert, H.J. | | Deposit date: | 2011-04-11 | | Release date: | 2011-05-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Novel, Noncatalytic Carbohydrate-Binding Module Displays Specificity for Galactose-Containing Polysaccharides Through Calcium-Mediated Oligomerization.
J.Biol.Chem., 286, 2011
|
|
3B5F
 
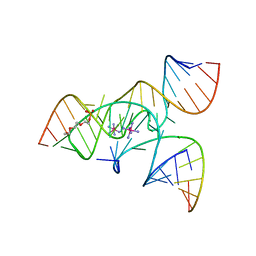 | | Crystal Structure of a Minimally Hinged Hairpin Ribozyme Incorporating the Ade38Dap Mutation and a 2',5' Phosphodiester Linkage at the Active Site | | Descriptor: | 29-mer Loop A and Loop B Ribozyme strand, COBALT HEXAMMINE(III), Loop A Substrate strand, ... | | Authors: | MacElrevey, C, Krucinska, J, Wedekind, J.E. | | Deposit date: | 2007-10-25 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural effects of nucleobase variations at key active site residue Ade38 in the hairpin ribozyme.
Rna, 14, 2008
|
|
1LWM
 
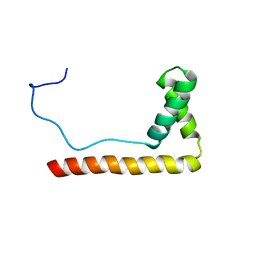 | | Solution Structure of the Sequence-Non-Specific HMGB protein NHP6A | | Descriptor: | NONHISTONE CHROMOSOMAL PROTEIN 6A | | Authors: | Masse, J.E, Wong, B, Yen, Y.-M, Allain, F.H.-T, Johnson, R.C, Feigon, J. | | Deposit date: | 2002-05-31 | | Release date: | 2002-10-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The S. cerevisiae architectural HMGB protein NHP6A complexed with DNA: DNA and protein conformational changes upon binding
J.Mol.Biol., 323, 2002
|
|
1LVM
 
 | | CATALYTICALLY ACTIVE TOBACCO ETCH VIRUS PROTEASE COMPLEXED WITH PRODUCT | | Descriptor: | CATALYTIC DOMAIN OF THE NUCLEAR INCLUSION PROTEIN A (NIA), OLIGOPEPTIDE SUBSTRATE FOR THE PROTEASE | | Authors: | Phan, J, Zdanov, A, Evdokimov, A.G, Tropea, J.E, Peters III, H.K, Kapust, R.B, Li, M, Wlodawer, A, Waugh, D.S. | | Deposit date: | 2002-05-28 | | Release date: | 2002-11-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the substrate specificity of tobacco etch virus protease.
J.Biol.Chem., 277, 2002
|
|
1MAZ
 
 | | X-RAY STRUCTURE OF BCL-XL, AN INHIBITOR OF PROGRAMMED CELL DEATH | | Descriptor: | Bcl-2-like protein 1 | | Authors: | Muchmore, S.W, Sattler, M, Liang, H, Meadows, R.P, Harlan, J.E, Yoon, H.S, Nettesheim, D, Chang, B.S, Thompson, C.B, Wong, S.L, Ng, S.C, Fesik, S.W. | | Deposit date: | 1996-04-09 | | Release date: | 1997-04-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray and NMR structure of human Bcl-xL, an inhibitor of programmed cell death.
Nature, 381, 1996
|
|
1MR2
 
 | | Structure of the MT-ADPRase in complex with 1 Mn2+ ion and AMP-CP (a inhibitor), a nudix enzyme | | Descriptor: | ADPR pyrophosphatase, MANGANESE (II) ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER | | Authors: | Kang, L.-W, Gabelli, S.B, Bianchet, M.A, Cunningham, J.E, O'Handley, S.F, Amzel, L.M. | | Deposit date: | 2002-09-17 | | Release date: | 2003-08-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and mechanism of MT-ADPRase, a Nudix hydrolase from Mycobacterium tuberculosis
Structure, 11, 2003
|
|
