1LVB
 
 | | CATALYTICALLY INACTIVE TOBACCO ETCH VIRUS PROTEASE COMPLEXED WITH SUBSTRATE | | Descriptor: | CATALYTIC DOMAIN OF THE NUCLEAR INCLUSION PROTEIN A (NIA), GLYCEROL, OLIGOPEPTIDE SUBSTRATE FOR THE PROTEASE | | Authors: | Phan, J, Zdanov, A, Evdokimov, A.G, Tropea, J.E, Peters III, H.K, Kapust, R.B, Li, M, Wlodawer, A, Waugh, D.S. | | Deposit date: | 2002-05-28 | | Release date: | 2002-11-27 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the substrate specificity of tobacco etch virus protease.
J.Biol.Chem., 277, 2002
|
|
2KXM
 
 | | Solution NMR Structure of the 27 nucleotide engineered neomycin sensing riboswitch RNA-ribostmycin complex | | Descriptor: | RIBOSTAMYCIN, RNA (27-MER) | | Authors: | Duchardt-Ferner, E, Weigand, J.E, Ohlenschlager, O, Schmidtke, S.R, Suess, B, Wohnert, J. | | Deposit date: | 2010-05-10 | | Release date: | 2011-04-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Highly modular structure and ligand binding by conformational capture in a minimalistic riboswitch.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
3BBK
 
 | | Miminally Junctioned Hairpin Ribozyme Incorporates A38C and 2'5'-phosphodiester Linkage within Active Site | | Descriptor: | COBALT HEXAMMINE(III), Loop A Substrate strand, Loop A and Loop B Ribozyme strand, ... | | Authors: | MacElrevey, C, Krucinska, J, Wedekind, J.E. | | Deposit date: | 2007-11-09 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural effects of nucleobase variations at key active site residue Ade38 in the hairpin ribozyme.
Rna, 14, 2008
|
|
3BBI
 
 | | Minimally Junctioned Hairpin Ribozyme Incorporating A38(2AP) and A-1 2'-O-Me Modifications near Active Site | | Descriptor: | COBALT HEXAMMINE(III), Loop A Substrate strand, Loop A and Loop B Ribozyme strand, ... | | Authors: | MacElrevey, C, Krucinska, J, Wedekind, J.E. | | Deposit date: | 2007-11-09 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural effects of nucleobase variations at key active site residue Ade38 in the hairpin ribozyme.
Rna, 14, 2008
|
|
3BBM
 
 | | Minimally Junctioned Hairpin Ribozyme Incorporates A38C and 2'O-Me Modification at Active Site | | Descriptor: | COBALT HEXAMMINE(III), Loop A Substrate strand, Loop A and Loop B Ribozyme strand, ... | | Authors: | MacElrevey, C, Krucinska, J, Wedekind, J.E. | | Deposit date: | 2007-11-09 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural effects of nucleobase variations at key active site residue Ade38 in the hairpin ribozyme.
Rna, 14, 2008
|
|
3BJI
 
 | | Structural Basis of Promiscuous Guanine Nucleotide Exchange by the T-Cell Essential Vav1 | | Descriptor: | Proto-oncogene vav, Ras-related C3 botulinum toxin substrate 1 precursor, ZINC ION | | Authors: | Chrencik, J.E, Brooun, A, Kuhn, P, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D) | | Deposit date: | 2007-12-04 | | Release date: | 2008-07-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of guanine nucleotide exchange mediated by the T-cell essential Vav1.
J.Mol.Biol., 380, 2008
|
|
3B7L
 
 | | Human farnesyl diphosphate synthase complexed with MG and minodronate | | Descriptor: | (1-HYDROXY-2-IMIDAZO[1,2-A]PYRIDIN-3-YLETHANE-1,1-DIYL)BIS(PHOSPHONIC ACID), Farnesyl pyrophosphate synthetase, MAGNESIUM ION | | Authors: | Pilka, E.S, Dunford, J.E, Guo, K, Pike, A.C.W, Kavanagh, K.L, von Delft, F, Ebetino, F.H, Arrowsmith, C.H, Edwards, A.M, Russell, R.G.G, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-10-31 | | Release date: | 2007-11-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Human farnesyl diphosphate synthase complexed with MG and minodronate.
TO BE PUBLISHED
|
|
2KX6
 
 | | Signaling state of Photoactive Yellow Protein | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Ramachandran, P.L, Lovett, J.E, Carl, P.J, Cammarata, M, Lee, J.H, Yang, J.O, Ihee, H, Timmel, C.R, van Thor, J. | | Deposit date: | 2010-04-27 | | Release date: | 2011-06-15 | | Last modified: | 2012-07-18 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | The short-lived signaling state of the photoactive yellow protein photoreceptor revealed by combined structural probes.
J.Am.Chem.Soc., 133, 2011
|
|
3B1U
 
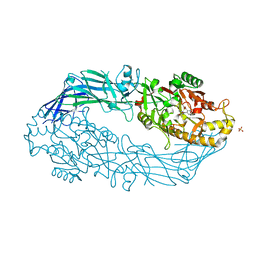 | | Crystal structure of human peptidylarginine deiminase 4 in complex with o-F-amidine | | Descriptor: | 2-{[(2S)-1-amino-5-{[(1Z)-2-fluoroethanimidoyl]amino}-1-oxopentan-2-yl]carbamoyl}benzoic acid, CALCIUM ION, Protein-arginine deiminase type-4, ... | | Authors: | Causey, C.P, Jones, J.E, Slack, J.L, Kamei, D, Jones Jr, L.E, Subramanian, V, Knuckley, B, Ebrahimi, P, Chumanevich, A.A, Luo, Y, Hashimoto, H, Shimizu, T, Sato, M, Hofseth, L.J, Thompson, P.R. | | Deposit date: | 2011-07-13 | | Release date: | 2011-10-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Development of N-alpha-(2-Carboxyl)benzoyl-N(5)-(2-fluoro-1-iminoethyl)-l-ornithine Amide (o-F-amidine) and N-alpha-(2-Carboxyl)benzoyl-N(5)-(2-chloro-1-iminoethyl)-l-ornithine Amide (o-Cl-amidine) As Second Generation Protein Arginine Deiminase (PAD) Inhibitors
J.Med.Chem., 54, 2011
|
|
3BGF
 
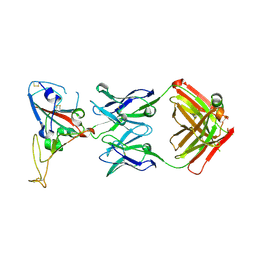 | |
2P4D
 
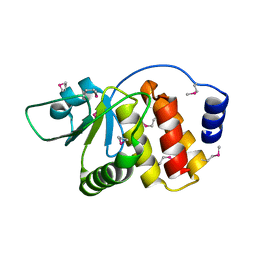 | |
3B91
 
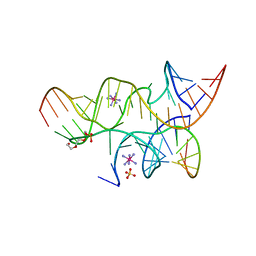 | | Minimally Hinged Hairpin Ribozyme Incorporates Ade38(2AP) and 2',5'-Phosphodiester Linkage Mutations at the Active Site | | Descriptor: | 29-mer Loop A and Loop B Ribozyme strand, COBALT HEXAMMINE(III), Loop A Substrate strand, ... | | Authors: | MacElrevey, C, Krucinska, J, Wedekind, J.E. | | Deposit date: | 2007-11-02 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural effects of nucleobase variations at key active site residue Ade38 in the hairpin ribozyme.
Rna, 14, 2008
|
|
3B58
 
 | | Minimally Junctioned Hairpin Ribozyme Incorporates A38G Mutation and a 2',5'-Phosphodiester Linkage at the Active Site | | Descriptor: | 29-mer Loop A and Loop B Ribozyme strand, COBALT HEXAMMINE(III), Loop A Substrate strand, ... | | Authors: | MacElrevey, C, Krucinska, J, Wedekind, J.E. | | Deposit date: | 2007-10-25 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural effects of nucleobase variations at key active site residue Ade38 in the hairpin ribozyme.
Rna, 14, 2008
|
|
5FL1
 
 | | Structure of a hydrolase with an inhibitor | | Descriptor: | (3~{a}~{R},5~{R},6~{S},7~{R},7~{a}~{R})-5-(hydroxymethyl)-2-(prop-2-enylamino)-5,6,7,7~{a}-tetrahydro-3~{a}~{H}-pyrano[3,2-d][1,3]thiazole-6,7-diol, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Cekic, N, Heinonen, J.E, Stubbs, K.A, Roth, C, McEachern, E.J, Davies, G.J, Vocadlo, D.J. | | Deposit date: | 2015-10-20 | | Release date: | 2016-08-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Analysis of transition state mimicry by tight binding aminothiazoline inhibitors provides insight into catalysis by humanO-GlcNAcase.
Chem Sci, 7, 2016
|
|
3ZIA
 
 | | The structure of F1-ATPase from Saccharomyces cerevisiae inhibited by its regulatory protein IF1 | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Robinson, G.C, Bason, J.V, Montgomery, M.G, Fearnley, I.M, Mueller, D.M, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2013-01-07 | | Release date: | 2013-02-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Structure of F1-ATPase from Saccharomyces Cerevisiae Inhibited by its Regulatory Protein If1.
Open Biol., 3, 2013
|
|
3ZTN
 
 | | STRUCTURE OF INFLUENZA A NEUTRALIZING ANTIBODY SELECTED FROM CULTURES OF SINGLE HUMAN PLASMA CELLS IN COMPLEX WITH HUMAN H1 INFLUENZA HAEMAGGLUTININ. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FI6V3 ANTIBODY LIGHT CHAIN, ... | | Authors: | Hubbard, P.A, Ritchie, A.J, Corti, D, Voss, J.E, Gamblin, S.J, Codoni, G, Macagno, A, Jarrossay, D, Pinna, D, Minola, A, Vanzetta, F, Silacci, C, Fernandez-Rodriguez, B.M, Agatic, G, Giacchetto-Sasselli, I, Vachieri, S.G, Sallusto, F, Collins, P.J, Haire, L.F, Temperton, N, Langedijk, J.P.M, Skehel, J.J, Lanzavecchia, A. | | Deposit date: | 2011-07-12 | | Release date: | 2011-08-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | A Neutralizing Antibody Selected from Plasma Cells that Binds to Group 1 and Group 2 Influenza a Hemagglutinins.
Science, 333, 2011
|
|
1GY9
 
 | | Taurine/alpha-ketoglutarate Dioxygenase from Escherichia coli | | Descriptor: | 2-AMINOETHANESULFONIC ACID, 2-OXOGLUTARIC ACID, ALPHA-KETOGLUTARATE-DEPENDENT TAURINE DIOXYGENASE, ... | | Authors: | Elkins, J.M, Burzlaff, N.I, Ryle, M.J, Lloyd, J.S, Clifton, I.J, Baldwin, J.E, Hausinger, R.P, Roach, P.L. | | Deposit date: | 2002-04-22 | | Release date: | 2002-04-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-Ray Crystal Structure of Escherichia Coli Taurine/Alpha-Ketoglutarate Dioxygenase Complexed to Ferrous Iron and Substrates.
Biochemistry, 41, 2002
|
|
1GNS
 
 | | SUBTILISIN BPN' | | Descriptor: | ACETONE, SUBTILISIN BPN' | | Authors: | Almog, O, Gallagher, D.T, Ladner, J.E, Strausberg, S, Alexander, P. | | Deposit date: | 2001-10-06 | | Release date: | 2002-06-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Thermostability. Analysis of Stabilizing Mutations in Subtilisin Bpn'.
J.Biol.Chem., 277, 2002
|
|
8TOA
 
 | | CryoEM structure of H7 hemagglutinin from A/Shanghai2/2013 H7N9 in complex with a human neutralizing antibody H7.HK2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, H7.HK2 Neutralizing Antibody Heavy Chain, H7.HK2 Neutralizing Antibody Light Chain, ... | | Authors: | Morano, N.C, Becker, J.E, Wu, X, Shapiro, L. | | Deposit date: | 2023-08-03 | | Release date: | 2024-05-15 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | CryoEM structure of H7 hemagglutinin from A/Shanghai2/2013 H7N9 in complex with a human neutralizing antibody H7.HK1
To Be Published
|
|
1H8H
 
 | | Bovine mitochondrial F1-ATPase crystallised in the presence of 5mm AMPPNP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, BOVINE MITOCHONDRIAL F1-ATPASE, ... | | Authors: | Braig, K, Menz, R.I, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2001-02-06 | | Release date: | 2001-04-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Structure and Nucleotide Occupancy of Bovine Mitochondrial F(1)-ATPase are not Influenced by Crystallisation at High Concentrations of Nucleotide
FEBS Lett., 494, 2001
|
|
3ZYS
 
 | | Human dynamin 1 deltaPRD polymer stabilized with GMPPCP | | Descriptor: | DYNAMIN-1, INTERFERON-INDUCED GTP-BINDING PROTEIN MX1 | | Authors: | Chappie, J.S, Mears, J.A, Fang, S, Leonard, M, Schmid, S.L, Milligan, R.A, Hinshaw, J.E, Dyda, F. | | Deposit date: | 2011-08-24 | | Release date: | 2011-10-12 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (12.2 Å) | | Cite: | A Pseudoatomic Model of the Dynamin Polymer Identifies a Hydrolysis-Dependent Powerstroke.
Cell(Cambridge,Mass.), 147, 2011
|
|
3ZWU
 
 | | Pseudomonas fluorescens PhoX in complex with vanadate, a transition state analogue | | Descriptor: | ALKALINE PHOSPHATASE PHOX, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yong, S.C, Roversi, P, Lillington, J.E.D, Zeldin, O.B, Garman, E.F, Lea, S.M, Berks, B.C. | | Deposit date: | 2011-08-02 | | Release date: | 2012-08-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | A Complex Iron-Calcium Cofactor Catalyzing Phosphotransfer Chemistry
Science, 345, 2014
|
|
5FV7
 
 | | Human Fen1 in complex with an N-hydroxyurea compound | | Descriptor: | 1-[(2S)-2,3-dihydro-1,4-benzodioxin-2-ylmethyl]-3-hydroxythieno[3,2-d]pyrimidine-2,4(1H,3H)-dione, FLAP ENDONUCLEASE 1, MAGNESIUM ION | | Authors: | Exell, J.C, Thompson, M.J, Finger, L.D, Shaw, S.K, Abbott, W.M, McWhirter, C, Debreczeni, J.E, Jones, C.D, Nissink, J.W.M, Ward, T.A, Sioberg, C.W.L, Molina, D.M, Durant, S.T, Grasby, J.A. | | Deposit date: | 2016-02-03 | | Release date: | 2016-08-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Cellular Active N-Hydroxyurea Fen1 Inhibitors Block Substrate Entry to the Active Site
Nat.Chem.Biol., 12, 2016
|
|
8TYM
 
 | |
1HB2
 
 | | ISOPENICILLIN N SYNTHASE FROM ASPERGILLUS NIDULANS (OXYGEN EXPOSED PRODUCT FROM ANAEROBIC ACOV FE COMPLEX) | | Descriptor: | FE (II) ION, ISOPENICILLIN N SYNTHASE, N6-[(1S)-2-{[(1R)-1-CARBOXY-2-METHYLPROPYL]OXY}-1-(MERCAPTOCARBONYL)-2-OXOETHYL]-6-OXO-L-LYSINE, ... | | Authors: | Ogle, J.M, Clifton, I.J, Rutledge, P.J, Elkins, J.M, Burzlaff, N.I, Adlington, R.M, Roach, P.L, Baldwin, J.E. | | Deposit date: | 2001-04-11 | | Release date: | 2001-11-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Alternative Oxidation by Isopenicillin N Synthase Observed by X-Ray Diffraction.
Chem.Biol., 8, 2001
|
|
