6P3S
 
 | |
6P3R
 
 | |
5TDQ
 
 | | Crystal Structure of the GOLD domain of ACBD3 | | Descriptor: | Golgi resident protein GCP60 | | Authors: | McPhail, J.A, Burke, J.E. | | Deposit date: | 2016-09-19 | | Release date: | 2016-11-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | The Molecular Basis of Aichi Virus 3A Protein Activation of Phosphatidylinositol 4 Kinase III beta , PI4KB, through ACBD3.
Structure, 25, 2017
|
|
7QJR
 
 | | Crystal structure of cutinase 1 from Thermobifida fusca DSM44342 (703) | | Descriptor: | Cutinase 1, TETRAETHYLENE GLYCOL | | Authors: | Zahn, M, Avilan, L, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Sourcing thermotolerant poly(ethylene terephthalate) hydrolase scaffolds from natural diversity
Nat Commun, 13, 2022
|
|
7QJO
 
 | |
7QJT
 
 | | Crystal structure of a cutinase enzyme from Thermobifida cellulosilytica TB100 (711) | | Descriptor: | GLYCEROL, MAGNESIUM ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Zahn, M, Shakespeare, T.J, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Sourcing thermotolerant poly(ethylene terephthalate) hydrolase scaffolds from natural diversity
Nat Commun, 13, 2022
|
|
4YG8
 
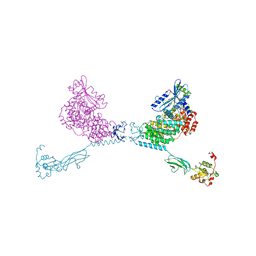 | | CRYSTAL STRUCTURE OF THE CHS5-CHS6 EXOMER CARGO ADAPTOR COMPLEX | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Chitin biosynthesis protein CHS5, Chitin biosynthesis protein CHS6, ... | | Authors: | Paczkowski, J.E, Richardson, B.C, Strassner, A.M, Fromme, J.C. | | Deposit date: | 2015-02-26 | | Release date: | 2015-03-11 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The exomer cargo adaptor structure reveals a novel GTPase-binding domain.
Embo J., 31, 2012
|
|
7QJQ
 
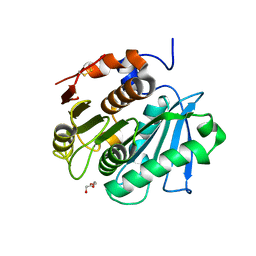 | | Crystal structure of a cutinase enzyme from Thermobifida fusca NTU22 (702) | | Descriptor: | Acetylxylan esterase, DI(HYDROXYETHYL)ETHER | | Authors: | Zahn, M, Gill, R.S, Avilan, L, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Sourcing thermotolerant poly(ethylene terephthalate) hydrolase scaffolds from natural diversity
Nat Commun, 13, 2022
|
|
3OK0
 
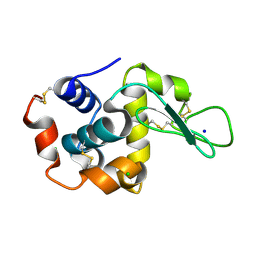 | | E35A Mutant of Hen Egg White Lysozyme (HEWL) | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | O'Meara, F, Bradley, J, O'Rourke, P.E, Webb, H, Tynan-Connolly, B.M, Nielsen, J.E. | | Deposit date: | 2010-08-24 | | Release date: | 2011-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | E35A Mutant of Hen Egg White Lysozyme (HEWL)
TO BE PUBLISHED
|
|
7QJP
 
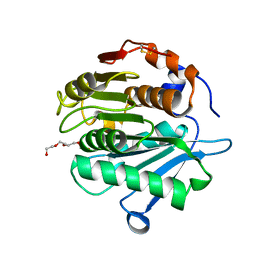 | | Crystal structure of a cutinase enzyme from Saccharopolyspora flava (611) | | Descriptor: | Cutinase, TETRAETHYLENE GLYCOL | | Authors: | Zahn, M, Avilan, L, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.561 Å) | | Cite: | Sourcing thermotolerant poly(ethylene terephthalate) hydrolase scaffolds from natural diversity
Nat Commun, 13, 2022
|
|
7QJM
 
 | |
7QJN
 
 | | Crystal structure of an alpha/beta-hydrolase enzyme from Candidatus Kryptobacter tengchongensis (306) | | Descriptor: | Dienelactone hydrolase, PHOSPHATE ION | | Authors: | Zahn, M, Gill, R.S, Erickson, E, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.885 Å) | | Cite: | Sourcing thermotolerant poly(ethylene terephthalate) hydrolase scaffolds from natural diversity
Nat Commun, 13, 2022
|
|
7QJS
 
 | | Crystal structure of a cutinase enzyme from Thermobifida fusca YX (705) | | Descriptor: | Cutinase 2, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Zahn, M, Shakespeare, T.J, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.429 Å) | | Cite: | Sourcing thermotolerant poly(ethylene terephthalate) hydrolase scaffolds from natural diversity
Nat Commun, 13, 2022
|
|
3OJP
 
 | | D52N Mutant of Hen Egg White Lysozyme (HEWL) | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | O'Meara, F, Bradley, J, O'Rourke, P.E, Webb, H, Tynan-Connolly, B.M, Nielsen, J.E. | | Deposit date: | 2010-08-23 | | Release date: | 2011-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | D52N Mutant of Hen Egg White Lysozyme (HEWL)
TO BE PUBLISHED
|
|
8G7L
 
 | | ATP-bound mtHsp60 V72I | | Descriptor: | 60 kDa heat shock protein, mitochondrial, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Braxton, J.R, Shao, H, Tse, E, Gestwicki, J.E, Southworth, D.R. | | Deposit date: | 2023-02-16 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Asymmetric apical domain states of mitochondrial Hsp60 coordinate substrate engagement and chaperonin assembly.
Biorxiv, 2023
|
|
8G7M
 
 | | ATP-bound mtHsp60 V72I focus | | Descriptor: | 60 kDa heat shock protein, mitochondrial, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Braxton, J.R, Shao, H, Tse, E, Gestwicki, J.E, Southworth, D.R. | | Deposit date: | 2023-02-16 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Asymmetric apical domain states of mitochondrial Hsp60 coordinate substrate engagement and chaperonin assembly.
Biorxiv, 2023
|
|
1SKY
 
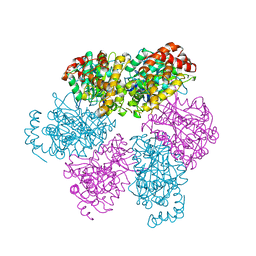 | | CRYSTAL STRUCTURE OF THE NUCLEOTIDE FREE ALPHA3BETA3 SUB-COMPLEX OF F1-ATPASE FROM THE THERMOPHILIC BACILLUS PS3 | | Descriptor: | F1-ATPASE, SULFATE ION | | Authors: | Shirakihara, Y, Leslie, A.G.W, Abrahams, J.P, Walker, J.E, Ueda, T, Sekimoto, Y, Kambara, M, Saika, K, Kagawa, Y, Yoshida, M. | | Deposit date: | 1997-02-26 | | Release date: | 1998-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The crystal structure of the nucleotide-free alpha 3 beta 3 subcomplex of F1-ATPase from the thermophilic Bacillus PS3 is a symmetric trimer.
Structure, 5, 1997
|
|
8G7O
 
 | | ATP- and mtHsp10-bound mtHsp60 V72I focus | | Descriptor: | 10 kDa heat shock protein, mitochondrial, 60 kDa heat shock protein, ... | | Authors: | Braxton, J.R, Shao, H, Tse, E, Gestwicki, J.E, Southworth, D.R. | | Deposit date: | 2023-02-16 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Asymmetric apical domain states of mitochondrial Hsp60 coordinate substrate engagement and chaperonin assembly.
Biorxiv, 2023
|
|
2A3K
 
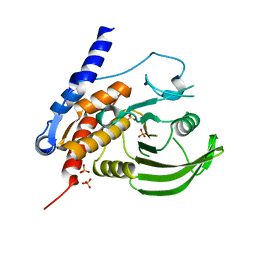 | | Crystal Structure of the Human Protein Tyrosine Phosphatase, PTPN7 (HePTP, Hematopoietic Protein Tyrosine Phosphatase) | | Descriptor: | PHOSPHATE ION, protein tyrosine phosphatase, non-receptor type 7, ... | | Authors: | Barr, A, Turnbull, A.P, Das, S, Eswaran, J, Debreczeni, J.E, Longmann, E, Smee, C, Burgess, N, Gileadi, O, Sundstrom, M, Arrowsmith, C, Edwards, A, von Delft, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-06-24 | | Release date: | 2005-07-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The crystal structure of human receptor protein tyrosine phosphatase kappa phosphatase domain 1.
Protein Sci., 15, 2006
|
|
5TIS
 
 | | Room temperature XFEL structure of the native, doubly-illuminated photosystem II complex | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Young, I.D, Ibrahim, M, Chatterjee, R, Gul, S, Fuller, F, Koroidov, S, Brewster, A.S, Tran, R, Alonso-Mori, R, Kroll, T, Michels-Clark, T, Laksmono, H, Sierra, R.G, Stan, C.A, Hussein, R, Zhang, M, Douthit, L, Kubin, M, de Lichtenberg, C, Pham, L.V, Nilsson, H, Cheah, M.H, Shevela, D, Saracini, C, Bean, M.A, Seuffert, I, Sokaras, D, Weng, T.-C, Pastor, E, Weninger, C, Fransson, T, Lassalle, L, Braeuer, P, Aller, P, Docker, P.T, Andi, B, Orville, A.M, Glownia, J.M, Nelson, S, Sikorski, M, Zhu, D, Hunter, M.S, Aquila, A, Koglin, J.E, Robinson, J, Liang, M, Boutet, S, Lyubimov, A.Y, Uervirojnangkoorn, M, Moriarty, N.W, Liebschner, D, Afonine, P.V, Watermann, D.G, Evans, G, Wernet, P, Dobbek, H, Weis, W.I, Brunger, A.T, Zwart, P.H, Adams, P.D, Zouni, A, Messinger, J, Bergmann, U, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2016-10-03 | | Release date: | 2016-11-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25000381 Å) | | Cite: | Structure of photosystem II and substrate binding at room temperature.
Nature, 540, 2016
|
|
8G7N
 
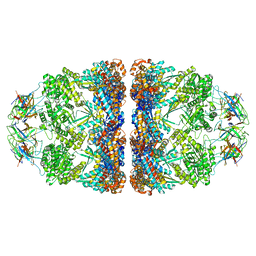 | | ATP- and mtHsp10-bound mtHsp60 V72I | | Descriptor: | 10 kDa heat shock protein, mitochondrial, 60 kDa heat shock protein, ... | | Authors: | Braxton, J.R, Shao, H, Tse, E, Gestwicki, J.E, Southworth, D.R. | | Deposit date: | 2023-02-16 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Asymmetric apical domain states of mitochondrial Hsp60 coordinate substrate engagement and chaperonin assembly.
Biorxiv, 2023
|
|
2D2V
 
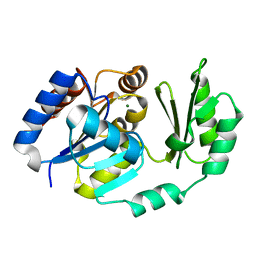 | |
4YCA
 
 | | Evidence of Kinetic Cooperativity in dimeric Ketopantoate Reductase from Staphylococcus aureus | | Descriptor: | 2-dehydropantoate 2-reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sanchez, J.E, Gross, P.G, Goetze, R, Walsh Jr, R.M, Peeples, W.B, Wood, Z.A. | | Deposit date: | 2015-02-19 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Evidence of Kinetic Cooperativity in Dimeric Ketopantoate Reductase from Staphylococcus aureus.
Biochemistry, 54, 2015
|
|
8G7K
 
 | | mtHsp60 V72I apo focus | | Descriptor: | 60 kDa heat shock protein, mitochondrial | | Authors: | Braxton, J.R, Shao, H, Tse, E, Gestwicki, J.E, Southworth, D.R. | | Deposit date: | 2023-02-16 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Asymmetric apical domain states of mitochondrial Hsp60 coordinate substrate engagement and chaperonin assembly.
Biorxiv, 2023
|
|
7QU9
 
 | | Structure of aminodeoxychorismate synthase component 1 (PabB) from Bacillus subtilis spizizenii. | | Descriptor: | Anthranilate synthase component I family protein, GLYCEROL, TRYPTOPHAN | | Authors: | Rooms, L.D, Race, P.R, Back, C.B, Burton, N.B, Willis, C.L, Stach, J.E.M, Duke, P.W, Hawkins, C. | | Deposit date: | 2022-01-17 | | Release date: | 2023-01-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structure of aminodeoxychorismate synthase component 1 (PabB) from Bacillus subtilis spizizenii.
To Be Published
|
|
