7M4H
 
 | | DNA Polymerase Lambda, dCTP:At Mn2+ Reaction State Ternary Complex, 225 min | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DNA (5'-D(*CP*AP*GP*TP*AP*CP*C)-3'), ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2021-03-21 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.923 Å) | | Cite: | Watching right and wrong nucleotide insertion captures hidden polymerase fidelity checkpoints.
Nat Commun, 13, 2022
|
|
7M49
 
 | | DNA Polymerase Lambda, TTP:At Mn2+ Reaction State Ternary Complex, 5 min | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*AP*GP*TP*AP*CP*T)-3'), DNA (5'-D(*CP*GP*GP*CP*AP*GP*TP*AP*CP*TP*G)-3'), ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2021-03-21 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Watching right and wrong nucleotide insertion captures hidden polymerase fidelity checkpoints.
Nat Commun, 13, 2022
|
|
7LAG
 
 | | CRYSTAL STRUCTURE OF MYELOPEROXIDASE SUBFORM C (MPO) COMPLEX WITH Compound-14 AKA 7-({1-[(3-phenoxyphenyl)methyl]-1H-pyrazol-4-yl}methyl)-3H-[1,2,3]triazolo[4,5-b]pyridin-5-amine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 7-({1-[(3-phenoxyphenyl)methyl]-1H-pyrazol-4-yl}methyl)-3H-[1,2,3]triazolo[4,5-b]pyridin-5-amine, ... | | Authors: | Khan, J.A. | | Deposit date: | 2021-01-06 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Small molecule and macrocyclic pyrazole derived inhibitors of myeloperoxidase (MPO).
Bioorg.Med.Chem.Lett., 42, 2021
|
|
7LAN
 
 | | CRYSTAL STRUCTURE OF MYELOPEROXIDASE SUBFORM C (MPO) COMPLEX WITH COMPOUND-30 AKA 7-[(3~{S},4~{R},6~{R})-4-benzyl-2-oxa-7,13,14-triazatetracyclo[14.3.1.1^{3,6}.1^{11,14}]docosa-1(19),11(21),12,16(20),17-pentaen-10-yl]-3~{H}-triazolo[4,5-b]pyridin-5-amine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 7-[(3R,4S,6S,10R)-4-benzyl-2-oxa-7,13,14-triazatetracyclo[14.3.1.1~3,6~.1~11,14~]docosa-1(20),11(21),12,16,18-pentaen-10-yl]-3H-[1,2,3]triazolo[4,5-b]pyridin-5-amine, ... | | Authors: | Khan, J.A. | | Deposit date: | 2021-01-06 | | Release date: | 2021-04-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Small molecule and macrocyclic pyrazole derived inhibitors of myeloperoxidase (MPO).
Bioorg.Med.Chem.Lett., 42, 2021
|
|
3SN6
 
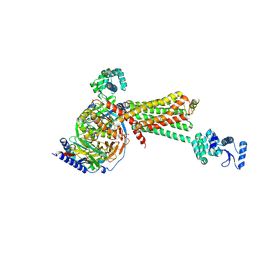 | | Crystal structure of the beta2 adrenergic receptor-Gs protein complex | | Descriptor: | 8-[(1R)-2-{[1,1-dimethyl-2-(2-methylphenyl)ethyl]amino}-1-hydroxyethyl]-5-hydroxy-2H-1,4-benzoxazin-3(4H)-one, Camelid antibody VHH fragment, Endolysin,Beta-2 adrenergic receptor, ... | | Authors: | Rasmussen, S.G.F, DeVree, B.T, Zou, Y, Kruse, A.C, Chung, K.Y, Kobilka, T.S, Thian, F.S, Chae, P.S, Pardon, E, Calinski, D, Mathiesen, J.M, Shah, S.T.A, Lyons, J.A, Caffrey, M, Gellman, S.H, Steyaert, J, Skiniotis, G, Weis, W.I, Sunahara, R.K, Kobilka, B.K. | | Deposit date: | 2011-06-28 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the beta2 adrenergic receptor-Gs protein complex
Nature, 477, 2011
|
|
3S3A
 
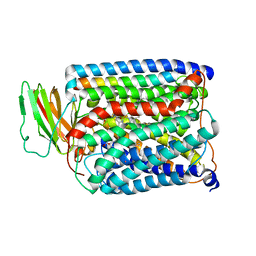 | | Structure of Thermus thermophilus cytochrome ba3 oxidase 120s after Xe depressurization | | Descriptor: | COPPER (II) ION, Cytochrome c oxidase polypeptide 2A, Cytochrome c oxidase subunit 1, ... | | Authors: | Luna, V.M, Fee, J.A, Deniz, A.A, Stout, C.D. | | Deposit date: | 2011-05-18 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (4.25 Å) | | Cite: | Mobility of Xe atoms within the oxygen diffusion channel of cytochrome ba(3) oxidase.
Biochemistry, 51, 2012
|
|
3S4T
 
 | | Crystal structure of putative amidohydrolase-2 (EFI-target 500288)from Polaromonas sp. JS666 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, Amidohydrolase 2, ... | | Authors: | Ramagopal, U.A, Toro, R, Girlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-05-20 | | Release date: | 2011-08-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of putative amidohydrolase-2 (EFI-target 500288)from Polaromonas sp. JS666
To be published
|
|
3S8F
 
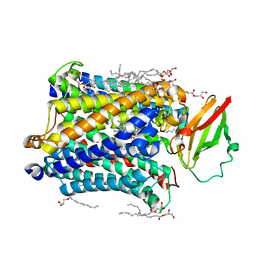 | | 1.8 A structure of ba3 cytochrome c oxidase from Thermus thermophilus in lipid environment | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, COPPER (II) ION, Cytochrome c oxidase polypeptide 2A, ... | | Authors: | Tiefenbrunn, T, Liu, W, Chen, Y, Katritch, V, Stout, C.D, Fee, J.A, Cherezov, V. | | Deposit date: | 2011-05-27 | | Release date: | 2011-08-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High resolution structure of the ba3 cytochrome c oxidase from Thermus thermophilus in a lipidic environment.
Plos One, 6, 2011
|
|
4R3R
 
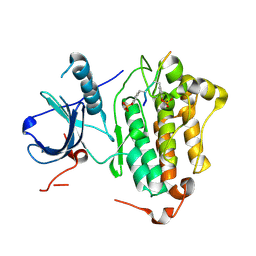 | | Crystal structures of EGFR in complex with Mig6 | | Descriptor: | Epidermal growth factor receptor, peptide from ERBB receptor feedback inhibitor 1' | | Authors: | Park, E, Kim, N, Yi, Z, Cho, A, Kim, K, Ficarro, S.B, Park, A, Park, W.Y, Murray, B, Meyerson, M, Beroukim, R, Marto, J.A, Cho, J, Eck, M.J. | | Deposit date: | 2014-08-17 | | Release date: | 2015-08-12 | | Last modified: | 2015-09-16 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure and mechanism of activity-based inhibition of the EGF receptor by Mig6.
Nat.Struct.Mol.Biol., 22, 2015
|
|
3S8E
 
 | |
7M4B
 
 | | DNA Polymerase Lambda, TTP:At Mn2+ Product State Ternary Complex, 60 min | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*AP*GP*TP*AP*CP*T)-3'), DNA (5'-D(*CP*GP*GP*CP*AP*GP*TP*AP*CP*TP*G)-3'), ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2021-03-21 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Watching right and wrong nucleotide insertion captures hidden polymerase fidelity checkpoints.
Nat Commun, 13, 2022
|
|
7M47
 
 | | DNA Polymerase Lambda, TTP:At Mg2+ Product State Ternary Complex, 60 min | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*AP*GP*TP*AP*CP*T)-3'), DNA (5'-D(*CP*GP*GP*CP*AP*GP*TP*AP*CP*TP*G)-3'), ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2021-03-21 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.648 Å) | | Cite: | Watching right and wrong nucleotide insertion captures hidden polymerase fidelity checkpoints.
Nat Commun, 13, 2022
|
|
7M43
 
 | | DNA Polymerase Lambda, TTP:At Ca2+ Ground State Ternary Complex | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DNA (5'-D(*CP*AP*GP*TP*AP*C)-3'), ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2021-03-21 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.978 Å) | | Cite: | Watching right and wrong nucleotide insertion captures hidden polymerase fidelity checkpoints.
Nat Commun, 13, 2022
|
|
4RBM
 
 | | Porphyromonas gingivalis gingipain K (Kgp) catalytic and immunoglobulin superfamily-like domains | | Descriptor: | (3S)-3,7-diaminoheptan-2-one, ACETATE ION, AZIDE ION, ... | | Authors: | de Diego, I, Veillard, F, Sztukowska, M.N, Guevara, T, Potempa, B, Pomowski, A, Huntington, J.A, Potempa, J, Gomis-Ruth, F.X. | | Deposit date: | 2014-09-12 | | Release date: | 2014-10-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and Mechanism of Cysteine Peptidase Gingipain K (Kgp), a Major Virulence Factor of Porphyromonas gingivalis in Periodontitis.
J.Biol.Chem., 289, 2014
|
|
7M4F
 
 | | DNA Polymerase Lambda, dCTP:At Mg2+ Product State Ternary Complex, 300 min | | Descriptor: | CHLORIDE ION, DNA (5'-D(*CP*AP*GP*TP*AP*CP*C)-3'), DNA (5'-D(*CP*GP*GP*CP*AP*GP*TP*AP*CP*TP*G)-3'), ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2021-03-21 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.947 Å) | | Cite: | Watching right and wrong nucleotide insertion captures hidden polymerase fidelity checkpoints.
Nat Commun, 13, 2022
|
|
7M4C
 
 | | DNA Polymerase Lambda, TTP:At Mn2+ Product State Ternary Complex, 960 min | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*AP*GP*TP*AP*CP*T)-3'), DNA (5'-D(*CP*GP*GP*CP*AP*GP*TP*AP*CP*TP*G)-3'), ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2021-03-21 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Watching right and wrong nucleotide insertion captures hidden polymerase fidelity checkpoints.
Nat Commun, 13, 2022
|
|
7M4J
 
 | | DNA Polymerase Lambda, dCTP:At Mn2+ Product State Ternary Complex, 960 min | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*AP*GP*TP*AP*CP*C)-3'), DNA (5'-D(*CP*GP*GP*CP*AP*GP*TP*AP*CP*TP*G)-3'), ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2021-03-21 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.377 Å) | | Cite: | Watching right and wrong nucleotide insertion captures hidden polymerase fidelity checkpoints.
Nat Commun, 13, 2022
|
|
7M4A
 
 | | DNA Polymerase Lambda, TTP:At Mn2+ Product State Ternary Complex, 20 min | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*AP*GP*TP*AP*CP*T)-3'), DNA (5'-D(*CP*GP*GP*CP*AP*GP*TP*AP*CP*TP*G)-3'), ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2021-03-21 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.868 Å) | | Cite: | Watching right and wrong nucleotide insertion captures hidden polymerase fidelity checkpoints.
Nat Commun, 13, 2022
|
|
7M4E
 
 | | DNA Polymerase Lambda, dCTP:At Mg2+ Reaction State Ternary Complex, 120 min | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DNA (5'-D(*CP*AP*GP*TP*AP*CP*C)-3'), ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2021-03-21 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Watching right and wrong nucleotide insertion captures hidden polymerase fidelity checkpoints.
Nat Commun, 13, 2022
|
|
7M46
 
 | | DNA Polymerase Lambda, TTP:At Mg2+ Product State Ternary Complex, 5 min | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*AP*GP*TP*AP*CP*T)-3'), DNA (5'-D(*CP*GP*GP*CP*AP*GP*TP*AP*CP*TP*G)-3'), ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2021-03-21 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Watching right and wrong nucleotide insertion captures hidden polymerase fidelity checkpoints.
Nat Commun, 13, 2022
|
|
7M44
 
 | | DNA Polymerase Lambda, TTP:At Mg2+ Reaction State Ternary Complex, 90 sec | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*AP*GP*TP*AP*CP*T)-3'), DNA (5'-D(*CP*GP*GP*CP*AP*GP*TP*AP*CP*TP*G)-3'), ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2021-03-21 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Watching right and wrong nucleotide insertion captures hidden polymerase fidelity checkpoints.
Nat Commun, 13, 2022
|
|
7M45
 
 | | DNA Polymerase Lambda, TTP:At Mg2+ Reaction State Ternary Complex, 120 sec | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*AP*GP*TP*AP*CP*T)-3'), DNA (5'-D(*CP*GP*GP*CP*AP*GP*TP*AP*CP*TP*G)-3'), ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2021-03-21 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.889 Å) | | Cite: | Watching right and wrong nucleotide insertion captures hidden polymerase fidelity checkpoints.
Nat Commun, 13, 2022
|
|
7M4K
 
 | | DNA Polymerase Lambda, TTPaS:At Ca2+ Ground State Ternary Complex | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DNA (5'-D(*CP*AP*GP*TP*AP*C)-3'), ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2021-03-21 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.719 Å) | | Cite: | Watching right and wrong nucleotide insertion captures hidden polymerase fidelity checkpoints.
Nat Commun, 13, 2022
|
|
7M4G
 
 | | DNA Polymerase Lambda, dCTP:At Mg2+ Product State Ternary Complex, 960 min | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*AP*GP*TP*AP*CP*C)-3'), DNA (5'-D(*CP*GP*GP*CP*AP*GP*TP*AP*CP*TP*G)-3'), ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2021-03-21 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Watching right and wrong nucleotide insertion captures hidden polymerase fidelity checkpoints.
Nat Commun, 13, 2022
|
|
7M4D
 
 | | DNA Polymerase Lambda, dCTP:At Ca2+ Ground State Ternary Complex | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2021-03-21 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.817 Å) | | Cite: | Watching right and wrong nucleotide insertion captures hidden polymerase fidelity checkpoints.
Nat Commun, 13, 2022
|
|
