7JX2
 
 | |
7JVG
 
 | | Cellular retinol-binding protein 2 (CRBP2) in complex with 1-arachidonoylglycerol | | Descriptor: | (2S)-2,3-dihydroxypropyl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate, Retinol-binding protein 2 | | Authors: | Silvaroli, J.A, Golczak, M. | | Deposit date: | 2020-08-21 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular basis for the interaction of cellular retinol binding protein 2 (CRBP2) with nonretinoid ligands.
J.Lipid Res., 62, 2021
|
|
7JVY
 
 | | Cellular retinol-binding protein 2 (CRBP2) in complex with 2-arachidonylglyceryl ether | | Descriptor: | 2-{[(5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraen-1-yl]oxy}propane-1,3-diol, Retinol-binding protein 2 | | Authors: | Silvaroli, J.A, Banarjee, S, Golczak, M. | | Deposit date: | 2020-08-24 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Molecular basis for the interaction of cellular retinol binding protein 2 (CRBP2) with nonretinoid ligands.
J.Lipid Res., 62, 2021
|
|
5ZO1
 
 | | Crystal structure of mouse nectin-like molecule 4 (mNecl-4) full ectodomain (Ig1-Ig3), 2.2A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Cell adhesion molecule 4, GLYCEROL | | Authors: | Liu, X, An, T, Li, D, Fan, Z, Xiang, P, Li, C, Ju, W, Li, J, Hu, G, Qin, B, Yin, B, Wojdyla, J.A, Wang, M, Yuan, J, Qiang, B, Shu, P, Cui, S, Peng, X. | | Deposit date: | 2018-04-12 | | Release date: | 2019-01-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structure of the heterophilic interaction between the nectin-like 4 and nectin-like 1 molecules.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5W1H
 
 | |
4CGN
 
 | | Leishmania major N-myristoyltransferase in complex with a piperidinylindole inhibitor | | Descriptor: | 2-(4-fluorophenyl)-N-(3-piperidin-4-yl-1H-indol-5-yl)ethanamide, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, MAGNESIUM ION, ... | | Authors: | Brannigan, J.A, Roberts, S.M, Bell, A.S, Hutton, J.A, Smith, D.F, Tate, E.W, Leatherbarrow, R.J, Wilkinson, A.J. | | Deposit date: | 2013-11-25 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Diverse Modes of Binding in Structures of Leishmania Major N-Myristoyltransferase with Selective Inhibitors
Iucrj, 1, 2014
|
|
5VTD
 
 | | Crystal Structure of the Co-bound Human Heavy-Chain Ferritin variant 122H-delta C-star | | Descriptor: | CALCIUM ION, CHLORIDE ION, COBALT (II) ION, ... | | Authors: | Bailey, J.B, Zhang, L, Chiong, J.A, Tezcan, F.A. | | Deposit date: | 2017-05-16 | | Release date: | 2017-06-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Synthetic Modularity of Protein-Metal-Organic Frameworks.
J. Am. Chem. Soc., 139, 2017
|
|
5W3V
 
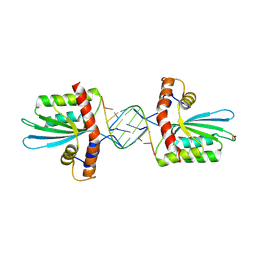 | | Crystal Structure of macaque APOBEC3H in complex with RNA | | Descriptor: | Apobec3H, RNA (5'-R(P*AP*AP*CP*CP*CP*CP*GP*GP*GP*C)-3'), RNA (5'-R(P*AP*AP*CP*CP*CP*GP*GP*GP*GP*A)-3'), ... | | Authors: | Bohn, J.A, Thummar, K, York, A, Raymond, A, Brown, W.C, Bieniasz, P.D, Hatziioannou, T, Smith, J.L. | | Deposit date: | 2017-06-08 | | Release date: | 2017-10-25 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.243 Å) | | Cite: | APOBEC3H structure reveals an unusual mechanism of interaction with duplex RNA.
Nat Commun, 8, 2017
|
|
5VJA
 
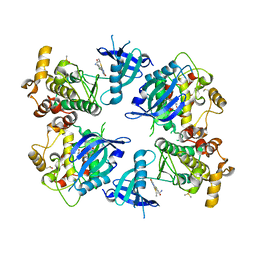 | | Crystal Structure of human zipper-interacting protein kinase (ZIPK, alias DAPK3) in complex with a pyrazolo[3,4-d]pyrimidinone ligand (HS38) | | Descriptor: | (2R)-2-{[1-(3-chlorophenyl)-4-oxo-4,5-dihydro-1H-pyrazolo[3,4-d]pyrimidin-6-yl]sulfanyl}propanamide, DIMETHYL SULFOXIDE, Death-associated protein kinase 3, ... | | Authors: | Carlson, D.A, Singer, M.R, Sutherland, C, Redondo, C, Alexander, L, Hughes, P.F, Knapp, S, MacDonald, J.A, Haystead, T.A.J. | | Deposit date: | 2017-04-19 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Targeting Pim Kinases and DAPK3 to Control Hypertension.
Cell Chem Biol, 25, 2018
|
|
5WHJ
 
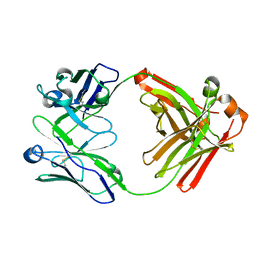 | | Crystal structure of Fab fragment of anti-FcRn antibody DX-2507 | | Descriptor: | DX-2507 Fab heavy chain, DX-2507 Fab light chain | | Authors: | Edwards, T.E, Fairman, J.W, Nixon, A.E, Kenniston, J.A. | | Deposit date: | 2017-07-17 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for pH-insensitive inhibition of immunoglobulin G recycling by an anti-neonatal Fc receptor antibody.
J. Biol. Chem., 292, 2017
|
|
7JVD
 
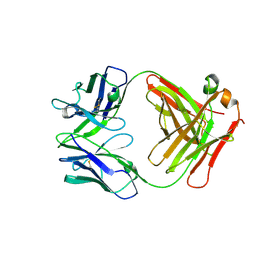 | | Fab of 5.6 monoclonal mouse IgG1 co-crystallized with the trisaccharide form of serotype 3 pneumococcal capsular polysaccharide | | Descriptor: | 5.6 Fab heavy chain, 5.6 Fab light chain, beta-D-glucopyranose-(1-3)-beta-D-glucopyranuronic acid-(1-4)-beta-D-glucopyranose | | Authors: | Ozdilek, A, Huang, J, Paschall, A.V, Babb, R, Middleton, D.R, Duke, J.A, Pirofski, L, Mousa, J.J, Avci, F.Y. | | Deposit date: | 2020-08-20 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Structural Model for the Ligand Binding of Pneumococcal Serotype 3 Capsular Polysaccharide-Specific Protective Antibodies.
Mbio, 12, 2021
|
|
7JT4
 
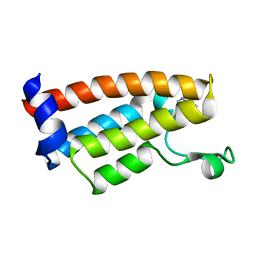 | | Crystal Structure of BPTF bromodomain labelled with 5-fluoro-tryptophan | | Descriptor: | Nucleosome-remodeling factor subunit BPTF | | Authors: | Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2020-08-17 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Selectivity, ligand deconstruction, and cellular activity analysis of a BPTF bromodomain inhibitor
Org.Biomol.Chem., 17, 2019
|
|
7JRS
 
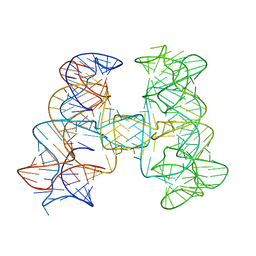 | |
7JRR
 
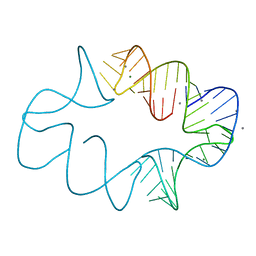 | | Crystal structures of artificially designed homomeric RNA nanoarchitectures | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, RNA (50-MER) | | Authors: | Liu, D, Shao, Y, Piccirilli, J.A, Weizmann, Y. | | Deposit date: | 2020-08-12 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structures of artificially designed discrete RNA nanoarchitectures at near-atomic resolution.
Sci Adv, 7, 2021
|
|
7JRT
 
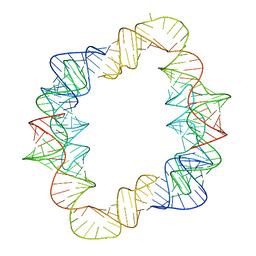 | |
7K06
 
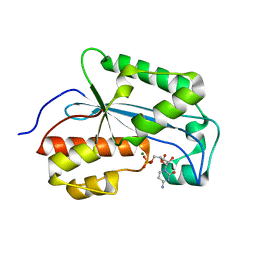 | |
7K07
 
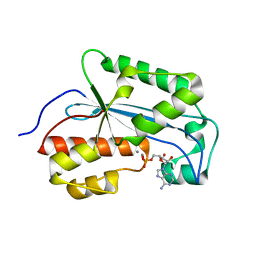 | |
7K05
 
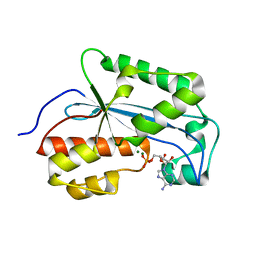 | |
5XGQ
 
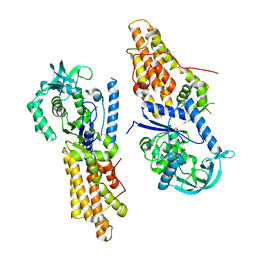 | |
2RIY
 
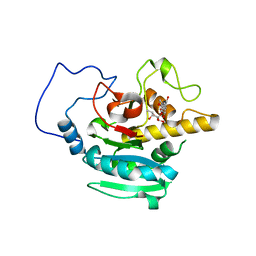 | | B-specific-1,3-galactosyltransferase (GTB)+H-antigen acceptor | | Descriptor: | Glycoprotein-fucosylgalactoside alpha-galactosyltransferase, octyl 2-O-(6-deoxy-alpha-L-galactopyranosyl)-beta-D-galactopyranoside | | Authors: | Evans, S.V, Alfaro, J.A. | | Deposit date: | 2007-10-13 | | Release date: | 2008-02-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | ABO(H) blood group A and B glycosyltransferases recognize substrate via specific conformational changes.
J.Biol.Chem., 283, 2008
|
|
2RJ4
 
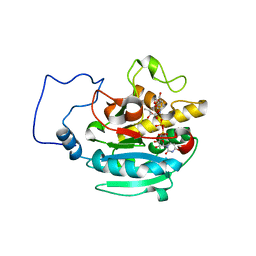 | | B-specific alpha-1,3-galactosyltransferase G176R +UDP+ADA | | Descriptor: | Glycoprotein-fucosylgalactoside alpha-galactosyltransferase, MANGANESE (II) ION, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Evans, S.V, Alfaro, J.A. | | Deposit date: | 2007-10-14 | | Release date: | 2008-02-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | ABO(H) blood group A and B glycosyltransferases recognize substrate via specific conformational changes.
J.Biol.Chem., 283, 2008
|
|
2RIX
 
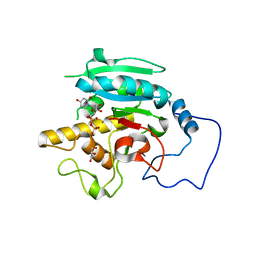 | | B-specific-1,3-galactosyltransferase)(GTB) + UDP | | Descriptor: | GLYCEROL, Glycoprotein-fucosylgalactoside alpha-galactosyltransferase, MANGANESE (II) ION, ... | | Authors: | Evans, S.V, Alfaro, J.A. | | Deposit date: | 2007-10-13 | | Release date: | 2008-02-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | ABO(H) blood group A and B glycosyltransferases recognize substrate via specific conformational changes.
J.Biol.Chem., 283, 2008
|
|
2RJ9
 
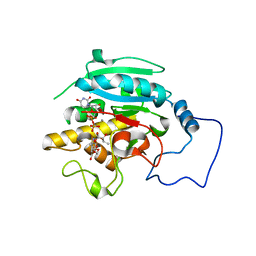 | |
8FE5
 
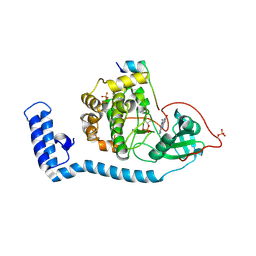 | | Structure of J-PKAc chimera complexed with Aplithianine B | | Descriptor: | 6-[(6P)-6-(1-methyl-1H-imidazol-5-yl)-2,3-dihydro-4H-1,4-thiazin-4-yl]-7,9-dihydro-8H-purin-8-one, DnaJ homolog subfamily B member 1,cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Du, L, Wilson, B.A.P, Li, N, Martinez Fiesco, J.A, Dalilian, M, Wang, D, Smith, E.A, Wamiru, A, Goncharova, E.I, Zhang, P, O'Keefe, B.R. | | Deposit date: | 2022-12-05 | | Release date: | 2023-10-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Discovery and Synthesis of a Naturally Derived Protein Kinase Inhibitor that Selectively Inhibits Distinct Classes of Serine/Threonine Kinases.
J.Nat.Prod., 86, 2023
|
|
7RAH
 
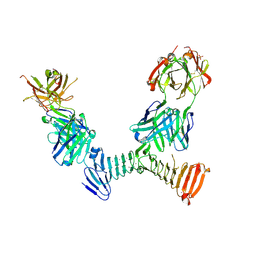 | | Adenylate cyclase toxin RTX domain fragment bound to M1H5 Fab and M2B10 Fab | | Descriptor: | Bifunctional adenylate cyclase toxin/hemolysin CyaA,Bifunctional adenylate cyclase toxin/hemolysin CyaA, CALCIUM ION, M1H5 Fab Heavy Chain, ... | | Authors: | Goldsmith, J.A, McLellan, J.S. | | Deposit date: | 2021-07-01 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for antibody binding to adenylate cyclase toxin reveals RTX linkers as neutralization-sensitive epitopes.
Plos Pathog., 17, 2021
|
|
