6TY6
 
 | | Variant W229D/F290W-2 of the last common ancestor of Gram-negative bacteria beta-lactamase class A (GNCA4) bound to 5(6)-nitrobenzotriazole (TS-analog) | | Descriptor: | 6-NITROBENZOTRIAZOLE, ACETATE ION, Beta lactamase (GNCA4-2), ... | | Authors: | Gavira, J.A, Risso, V, Sanchez-Ruiz, J.M, Romero-Rivera, A, Kamerlin, S.C.L, Ortega-Munoz, M, Santoyo-Gonzalez, F. | | Deposit date: | 2020-01-15 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Enhancing ade novoenzyme activity by computationally-focused ultra-low-throughput screening.
Chem Sci, 11, 2020
|
|
6U9M
 
 | | MLL1 SET N3861I/Q3867L bound to inhibitor 16 (TC-5109) | | Descriptor: | 5'-{[(3S)-3-amino-3-carboxypropyl]({1-[(thiophen-2-yl)methyl]azetidin-3-yl}methyl)amino}-5'-deoxyadenosine, GLYCEROL, Histone-lysine N-methyltransferase, ... | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2019-09-09 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of Potent Small-Molecule Inhibitors of MLL Methyltransferase.
Acs Med.Chem.Lett., 11, 2020
|
|
6U8D
 
 | | Crystal structure of hepatitis C virus IRES junction IIIabc in complex with Fab HCV2 | | Descriptor: | Heavy chain of Fab HCV2, JIIIabc RNA (68-MER), Light chain of Fab HCV2 | | Authors: | Koirala, D, Lewicka, A, Koldobskaya, Y, Huang, H, Piccirilli, J.A. | | Deposit date: | 2019-09-04 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.807 Å) | | Cite: | Synthetic Antibody Binding to a Preorganized RNA Domain of Hepatitis C Virus Internal Ribosome Entry Site Inhibits Translation.
Acs Chem.Biol., 15, 2020
|
|
5O4V
 
 | | P.vivax NMT with aminomethylindazole and quinoline inhibitors bound | | Descriptor: | 1-[5-(4-fluoranyl-2-methyl-phenyl)-1~{H}-indazol-3-yl]-~{N},~{N}-dimethyl-methanamine, 2-oxopentadecyl-CoA, CHLORIDE ION, ... | | Authors: | Brannigan, J.A, Wilkinson, A.J. | | Deposit date: | 2017-05-31 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Fragment-derived inhibitors of human N-myristoyltransferase block capsid assembly and replication of the common cold virus.
Nat Chem, 10, 2018
|
|
7R9V
 
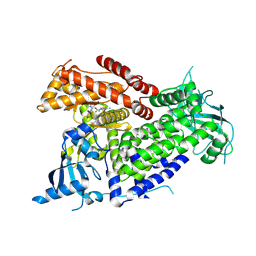 | | Structure of PIK3CA with covalent inhibitor 19 | | Descriptor: | N-[2-(4-{4-[2-amino-4-(difluoromethyl)pyrimidin-5-yl]-6-(morpholin-4-yl)-1,3,5-triazin-2-yl}piperazin-1-yl)-2-oxoethyl]-1-(prop-2-enoyl)piperidine-4-carboxamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Burke, J.E, McPhail, J.A. | | Deposit date: | 2021-06-29 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Covalent Proximity Scanning of a Distal Cysteine to Target PI3K alpha.
J.Am.Chem.Soc., 144, 2022
|
|
7R9Y
 
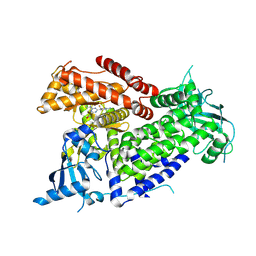 | | Structure of PIK3CA with covalent inhibitor 22 | | Descriptor: | N-[2-(4-{4-[2-amino-4-(difluoromethyl)pyrimidin-5-yl]-6-(morpholin-4-yl)-1,3,5-triazin-2-yl}piperazin-1-yl)-2-oxoethyl]-N-methyl-1-(prop-2-enoyl)piperidine-4-carboxamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Burke, J.E, McPhail, J.A. | | Deposit date: | 2021-06-29 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Covalent Proximity Scanning of a Distal Cysteine to Target PI3K alpha.
J.Am.Chem.Soc., 144, 2022
|
|
7R51
 
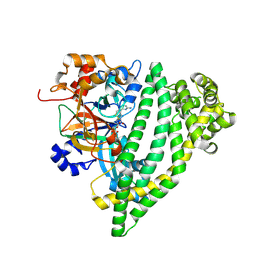 | | Structure of P. gingivalis DPP11 in complex with the dipeptide Arg-Asp | | Descriptor: | ARG-ASP, Asp/Glu-specific dipeptidyl-peptidase | | Authors: | Tham, C.T, Coker, J.A, Foster, W.R, Ohara-Nemoto, Y, Nemoto, T.K, Yue, W.W, Bountra, C, Bezerra, G.A. | | Deposit date: | 2022-02-09 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.808 Å) | | Cite: | Structure of P. gingivalis DPP11 in complex with the dipeptide Arg-Asp
To Be Published
|
|
6UOP
 
 | |
6UOR
 
 | |
6UOU
 
 | |
6UUR
 
 | | Human prion protein fibril, M129 variant | | Descriptor: | Major prion protein | | Authors: | Glynn, C, Sawaya, M.R, Ge, P, Zhou, Z.H, Rodriguez, J.A. | | Deposit date: | 2019-10-31 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of a human prion fibril with a hydrophobic, protease-resistant core.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6UVJ
 
 | |
6UOQ
 
 | |
7R0T
 
 | | Crystal structure of exonuclease ExnV1 | | Descriptor: | CHLORIDE ION, Exonuclease ExnV1, MAGNESIUM ION, ... | | Authors: | Welin, M, Svensson, A, Hakansson, M, Al-Karadaghi, S, Jasilionis, A, Linares-Pasten, J.A, Wang, L, Nordberg Karlsson, E, Ahlqvist, J. | | Deposit date: | 2022-02-02 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | Crystal structure of DNA polymerase I from Thermus phage G20c.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7R0K
 
 | | Crystal structure of Polymerase I from phage G20c | | Descriptor: | DNA polymerase I | | Authors: | Welin, M, Svensson, A, Hakansson, M, Al-Karadaghi, S, Linares-Pasten, J.A, Jasilionis, A, Nordberg Karlsson, E, Ahlqvist, J. | | Deposit date: | 2022-02-02 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.972 Å) | | Cite: | Crystal structure of DNA polymerase I from Thermus phage G20c.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7RRN
 
 | |
6UPS
 
 | |
6UPU
 
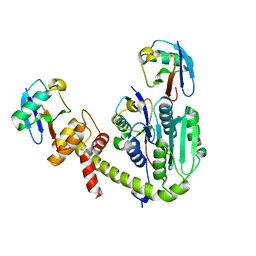 | |
7S65
 
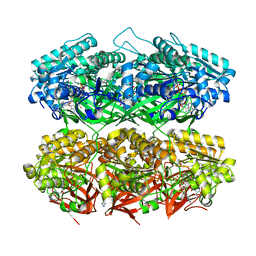 | | Compressed conformation of nighttime state KaiC | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein kinase KaiC, ... | | Authors: | Sandate, C.R, Swan, J.A, Partch, C.L, Lander, G.C. | | Deposit date: | 2021-09-13 | | Release date: | 2021-09-22 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Coupling of distant ATPase domains in the circadian clock protein KaiC.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7S67
 
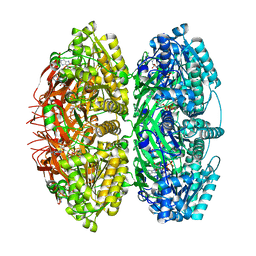 | | Extended conformation of daytime state KaiC | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein kinase KaiC, ... | | Authors: | Sandate, C.R, Swan, J.A, Partch, C.L, Lander, G.C. | | Deposit date: | 2021-09-13 | | Release date: | 2021-09-22 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Coupling of distant ATPase domains in the circadian clock protein KaiC.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7S66
 
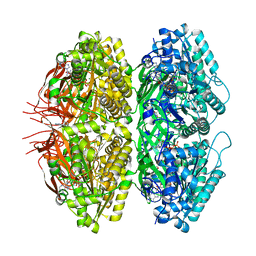 | | Extended conformation of nighttime state KaiC | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein kinase KaiC, MAGNESIUM ION | | Authors: | Sandate, C.R, Swan, J.A, Partch, C.L, Lander, G.C. | | Deposit date: | 2021-09-13 | | Release date: | 2021-09-22 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Coupling of distant ATPase domains in the circadian clock protein KaiC.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RWT
 
 | | Adeno-associated virus type 2 | | Descriptor: | Capsid protein VP1 | | Authors: | Hull, J.A, Mietzsch, M, Chipman, P, Strugatsky, D, McKenna, R. | | Deposit date: | 2021-08-20 | | Release date: | 2021-10-20 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | Structural characterization of an envelope-associated adeno-associated virus type 2 capsid.
Virology, 565, 2021
|
|
7RWL
 
 | | Envelope-associated Adeno-associated virus serotype 2 | | Descriptor: | Capsid protein VP1 | | Authors: | Hull, J.A, Mietzsch, M, Chipman, P, Strugatsky, D, McKenna, R. | | Deposit date: | 2021-08-20 | | Release date: | 2021-10-20 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structural characterization of an envelope-associated adeno-associated virus type 2 capsid.
Virology, 565, 2021
|
|
7S3N
 
 | |
7S3M
 
 | |
