2XIC
 
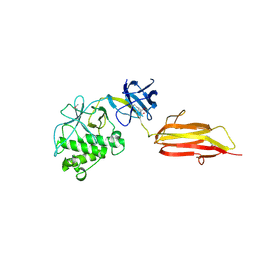 | | Pilus-presented adhesin, Spy0125 (Cpa), P212121 form (ESRF data) | | Descriptor: | ANCILLARY PROTEIN 1 | | Authors: | Pointon, J.A, Smith, W.D, Saalbach, G, Crow, A, Kehoe, M.A, Banfield, M.J. | | Deposit date: | 2010-06-28 | | Release date: | 2010-08-04 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A Highly Unusual Thioester Bond in a Pilus Adhesin is Required for Efficient Host Cell Interaction
J.Biol.Chem., 285, 2010
|
|
2XCL
 
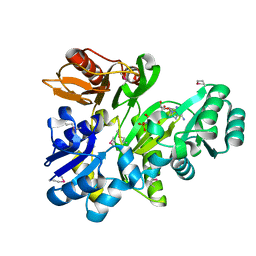 | |
6UFH
 
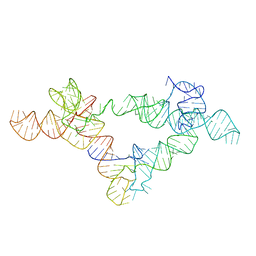 | |
2XM3
 
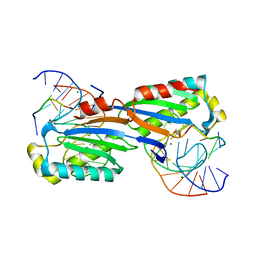 | | Deinococcus radiodurans ISDra2 Transposase Left end DNA complex | | Descriptor: | 5'-D(*TP*TP*AP*GP*T)-3', ACETATE ION, DRA2 TRANSPOSASE BINDING ELEMENT, ... | | Authors: | Hickman, A.B, James, J.A, Barabas, O, Pasternak, C, Ton-Hoang, B, Chandler, M, Sommer, S, Dyda, F. | | Deposit date: | 2010-07-22 | | Release date: | 2010-10-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA Recognition and the Precleavage State During Single-Stranded DNA Transposition in D. Radiodurans.
Embo J., 29, 2010
|
|
4AXS
 
 | | Structure of Carbamate Kinase from Mycoplasma penetrans | | Descriptor: | CARBAMATE KINASE, SULFATE ION | | Authors: | Gallego, P, Planell, R, Benach, J, Querol, E, PerezPons, J.A, Reverter, D. | | Deposit date: | 2012-06-14 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Characterization of the Enzymes Composing the Arginine Deiminase Pathway in Mycoplasma Penetrans.
Plos One, 7, 2012
|
|
2XLL
 
 | | The crystal structure of bilirubin oxidase from Myrothecium verrucaria | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIRUBIN OXIDASE, COPPER (II) ION | | Authors: | McNamara, T.P, Lowe, E.D, Cracknell, J.A, Blanford, C.F. | | Deposit date: | 2010-07-21 | | Release date: | 2011-04-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.305 Å) | | Cite: | Bilirubin Oxidase from Myrothecium Verrucaria: X- Ray Determination of the Complete Crystal Structure and a Rational Surface Modification for Enhanced Electrocatalytic O(2) Reduction.
Dalton Trans, 40, 2011
|
|
1O90
 
 | | Methionine Adenosyltransferase complexed with a L-methionine analogue | | Descriptor: | (2S,4S)-2-AMINO-4,5-EPOXIPENTANOIC ACID, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Gonzalez, B, Pajares, M.A, Hermoso, J.A, Sanz-Aparicio, J. | | Deposit date: | 2002-12-10 | | Release date: | 2003-08-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structures of Methionine Adenosyltransferase Complexed with Substrates and Products Reveal the Methionine-ATP Recognition and Give Insights Into the Catalytic Mechanism
J.Mol.Biol., 331, 2003
|
|
1WB6
 
 | | S954A mutant of the feruloyl esterase module from clostridium thermocellum complexed with vanillate | | Descriptor: | ACETATE ION, CADMIUM ION, ENDO-1,4-BETA-XYLANASE Y, ... | | Authors: | Tarbouriech, N, Prates, J.A, Fontes, C, Davies, G.J. | | Deposit date: | 2004-10-30 | | Release date: | 2006-05-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular Determinants of Substrate Specificity in the Feruloyl Esterase Module of Xylanase 10B from Clostridium Thermocellum
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2YN1
 
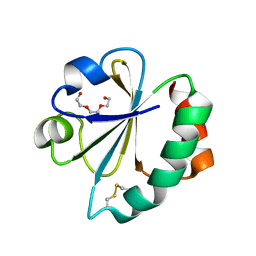 | | Crystal Structure of Ancestral Thioredoxin Relative to Last Gamma- Proteobacteria Common Ancestor (LGPCA) from the Precambrian Period | | Descriptor: | LGPCA THIOREDOXIN, TRIETHYLENE GLYCOL | | Authors: | Gavira, J.A, Ingles-Prieto, A, Ibarra-Molero, B, Sanchez-Ruiz, J.M. | | Deposit date: | 2012-10-11 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Conservation of Protein Structure Over Four Billion Years
Structure, 21, 2013
|
|
1O92
 
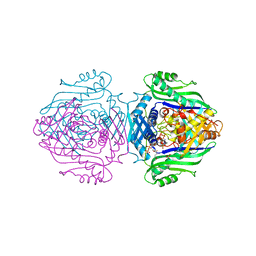 | | Methionine Adenosyltransferase complexed with ADP and a L-methionine analogue | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, L-2-AMINO-4-METHOXY-CIS-BUT-3-ENOIC ACID, MAGNESIUM ION, ... | | Authors: | Gonzalez, B, Pajares, M.A, Hermoso, J.A, Sanz-Aparicio, J. | | Deposit date: | 2002-12-10 | | Release date: | 2003-08-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Crystal Structures of Methionine Adenosyltransferase Complexed with Substrates and Products Reveal the Methionine-ATP Recognition and Give Insights Into the Catalytic Mechanism
J.Mol.Biol., 331, 2003
|
|
1O9T
 
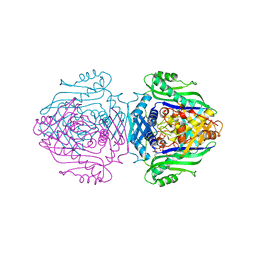 | | Methionine adenosyltransferase complexed with both substrates ATP and methionine | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, METHIONINE, ... | | Authors: | Gonzalez, B, Pajares, M.A, Hermoso, J.A, Sanz-Aparicio, J. | | Deposit date: | 2002-12-18 | | Release date: | 2003-08-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structures of Methionine Adenosyltransferase Complexed with Substrates and Products Reveal the Methionine-ATP Recognition and Give Insights Into the Catalytic Mechanism
J.Mol.Biol., 331, 2003
|
|
1VYK
 
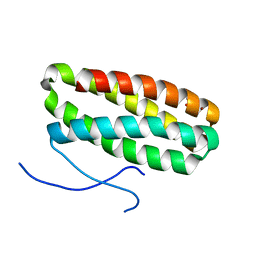 | | CRYSTAL STRUCTURE OF PSBQ PROTEIN OF PHOTOSYSTEM II FROM HIGHER PLANTS | | Descriptor: | ACETATE ION, OXYGEN-EVOLVING ENHANCER PROTEIN 3, ZINC ION | | Authors: | Hermoso, J.A, Balsera, M, De Las Rivas, J, Arellano, J.B. | | Deposit date: | 2004-04-30 | | Release date: | 2005-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | The 1.49 A Resolution Crystal Structure of Psbq from Photosystem II of Spinacia Oleracea Reveals a Ppii Structure in the N-Terminal Region.
J.Mol.Biol., 350, 2005
|
|
2YNE
 
 | | Plasmodium vivax N-myristoyltransferase in complex with a benzothiophene inhibitor | | Descriptor: | 2-(3-piperidin-4-yloxy-1-benzothiophen-2-yl)-5-[(1,3,5-trimethylpyrazol-4-yl)methyl]-1,3,4-oxadiazole, 2-oxopentadecyl-CoA, CHLORIDE ION, ... | | Authors: | Wright, M.H, Clough, B, Rackham, M.D, Brannigan, J.A, Grainger, M, Bottrill, A.R, Heal, W.P, Broncel, M, Serwa, R.A, Mann, D, Leatherbarrow, R.J, Wilkinson, A.J, Holder, A.A, Tate, E.W. | | Deposit date: | 2012-10-13 | | Release date: | 2014-01-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Validation of N-Myristoyltransferase as an Antimalarial Drug Target Using an Integrated Chemical Biology Approach.
Nat.Chem., 6, 2014
|
|
1W35
 
 | | FERREDOXIN-NADP+ REDUCTASE (MUTATION: Y 303 W) | | Descriptor: | FERREDOXIN-NADP+ REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Hermoso, J.A, Perez-Dorado, I, Medina, M, Julvez, M.M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 2004-07-13 | | Release date: | 2005-07-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | C-Terminal Tyrosine of Ferredoxin-Nadp(+) Reductase in Hydride Transfer Processes with Nad(P)(+)/H.
Biochemistry, 44, 2005
|
|
1W34
 
 | | FERREDOXIN-NADP REDUCTASE (MUTATION: Y 303 S) | | Descriptor: | FERREDOXIN-NADP REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Hermoso, J.A, Perez-Dorado, I, Medina, M, Julvez, M.M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 2004-07-13 | | Release date: | 2005-10-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | C-Terminal Tyrosine of Ferredoxin-Nadp(+) Reductase in Hydride Transfer Processes with Nad(P)(+)/H.
Biochemistry, 44, 2005
|
|
1W52
 
 | | Crystal structure of a proteolyzed form of pancreatic lipase related protein 2 from horse | | Descriptor: | CALCIUM ION, DECYLAMINE-N,N-DIMETHYL-N-OXIDE, PANCREATIC LIPASE RELATED PROTEIN 2 | | Authors: | Mancheno, J.M, Jayne, S, Kerfelec, B, Chapus, C, Crenon, I, Hermoso, J.A. | | Deposit date: | 2004-08-04 | | Release date: | 2006-07-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Crystalization of a Proteolyzed Form of the Horse Pancreatic Lipase-Related Protein 2: Structural Basis for the Specific Detergent Requirement.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1O93
 
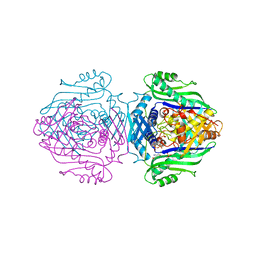 | | Methionine Adenosyltransferase complexed with ATP and a L-methionine analogue | | Descriptor: | (2S,4S)-2-AMINO-4,5-EPOXIPENTANOIC ACID, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Gonzalez, B, Pajares, M.A, Hermoso, J.A, Sanz-Aparicio, J. | | Deposit date: | 2002-12-10 | | Release date: | 2003-08-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Crystal Structures of Methionine Adenosyltransferase Complexed with Substrates and Products Reveal the Methionine-ATP Recognition and Give Insights Into the Catalytic Mechanism
J.Mol.Biol., 331, 2003
|
|
1WO6
 
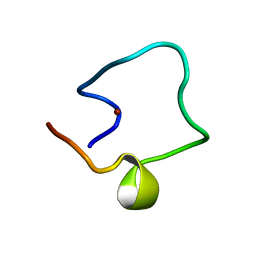 | | Solution structure of Designed Functional Finger 5 (DFF5): Designed mutant based on non-native CHANCE domain | | Descriptor: | CREB Binding Protein, ZINC ION | | Authors: | Sharpe, B.K, Liew, C.K, Wilce, J.A, Crossley, M, Matthews, J.M, Mackay, J.P. | | Deposit date: | 2004-08-12 | | Release date: | 2005-03-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Assessment of the robustness of a serendipitous zinc binding fold: mutagenesis and protein grafting
Structure, 13, 2005
|
|
1WVN
 
 | |
2YN4
 
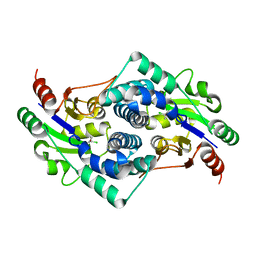 | | L-2-chlorobutryic acid bound complex L-haloacid dehalogenase from a Rhodobacteraceae family bacterium | | Descriptor: | (2S)-2-chlorobutanoic acid, L-HALOACID DEHALOGENASE | | Authors: | Novak, H.R, Sayer, C, Isupov, M.N, Paszkiewicz, K, Gotz, D, Spragg, A.M, Littlechild, J.A. | | Deposit date: | 2012-10-12 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Marine Rhodobacteraceae L-Haloacid Dehalogenase Contains a Novel His/Glu Dyad that Could Activate the Catalytic Water.
FEBS J., 280, 2013
|
|
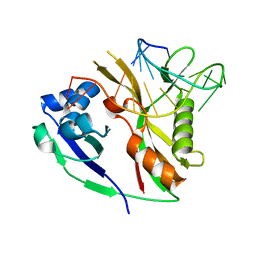
2Y8Y
 
 | |
2Y8W
 
 | |
2YEV
 
 | | Structure of caa3-type cytochrome oxidase | | Descriptor: | (1R,4S,6R)-6-({[2-(ACETYLAMINO)-2-DEOXY-ALPHA-D-GLUCOPYRANOSYL]OXY}METHYL)-4-HYDROXY-1-{[(15-METHYLHEXADECANOYL)OXY]METHYL}-4-OXIDO-7-OXO-3,5-DIOXA-8-AZA-4-PHOSPHAHEPTACOS-1-YL 15-METHYLHEXADECANOATE, (2R)-2,3-DIHYDROXYPROPYL (7Z)-TETRADEC-7-ENOATE, (2R)-3-HYDROXYPROPANE-1,2-DIYL DIHEXADECANOATE, ... | | Authors: | Lyons, J.A, Aragao, D, Soulimane, T, Caffrey, M. | | Deposit date: | 2011-03-31 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural Insights Into Electron Transfer in Caa3-Type Cytochrome Oxidases.
Nature, 487, 2012
|
|
2Y9H
 
 | |
2VR8
 
 | | Crystal Structure of G85R ALS mutant of Human Cu,Zn Superoxide Dismutase (CuZnSOD) at 1.36 A resolution | | Descriptor: | COPPER (II) ION, SULFATE ION, SUPEROXIDE DISMUTASE [CU-ZN], ... | | Authors: | Antonyuk, S, Cao, X, Seetharaman, S.V, Whitson, L.J, Taylor, A.B, Holloway, S.P, Strange, R.W, Doucette, P.A, Tiwari, A, Hayward, L.J, Padua, S, Cohlberg, J.A, Selverstone Valentine, J, Hasnain, S.S, Hart, P.J. | | Deposit date: | 2008-03-28 | | Release date: | 2008-04-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structures of the G85R Variant of Sod1 in Familial Amyotrophic Lateral Sclerosis.
J.Biol.Chem., 283, 2008
|
|
