2VTN
 
 | | Identification of N-(4-piperidinyl)-4-(2,6-dichlorobenzoylamino)-1H- pyrazole-3-carboxamide (AT7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design. | | Descriptor: | 4-(acetylamino)-N-(4-fluorophenyl)-1H-pyrazole-3-carboxamide, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Wyatt, P.G, Woodhead, A.J, Boulstridge, J.A, Berdini, V, Carr, M.G, Cross, D.M, Danillon, D, Davis, D.J, Devine, L.A, Early, T.R, Feltell, R.E, Lewis, E.J, McMenamin, R.L, Navarro, E.F, O'Brien, M.A, O'Reilly, M, Reule, M, Saxty, G, Seavers, L.C.A, Smith, D, Squires, M.S, Trewartha, G, Walker, M.T, Woolford, A.J. | | Deposit date: | 2008-05-15 | | Release date: | 2008-08-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of N-(4-Piperidinyl)-4-(2,6-Dichlorobenzoylamino)-1H-Pyrazole-3-Carboxamide (at7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design.
J.Med.Chem., 51, 2008
|
|
2W17
 
 | | CDK2 in complex with the imidazole pyrimidine amide, compound (S)-8b | | Descriptor: | ACETATE ION, CELL DIVISION PROTEIN KINASE 2, N-(4-{[(3S)-3-(dimethylamino)pyrrolidin-1-yl]carbonyl}phenyl)-5-fluoro-4-[2-methyl-1-(1-methylethyl)-1H-imidazol-5-yl]pyrimidin-2-amine | | Authors: | Jones, C.D, Andrews, D.M, Barker, A.J, Blades, K, Daunt, P, East, S, Geh, C, Graham, M.A, Johnson, K.M, Loddick, S.A, McFarland, H.M, McGregor, A, Moss, L, Rudge, D.A, Simpson, P.B, Swain, M.L, Tam, K.Y, Tucker, J.A, Walker, M, Brassington, C, Haye, H, McCall, E. | | Deposit date: | 2008-10-15 | | Release date: | 2008-11-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Discovery of Azd5597, a Potent Imidazole Pyrimidine Amide Cdk Inhibitor Suitable for Intravenous Dosing.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2VTQ
 
 | | Identification of N-(4-piperidinyl)-4-(2,6-dichlorobenzoylamino)-1H- pyrazole-3-carboxamide (AT7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design. | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, {[(2,6-difluorophenyl)carbonyl]amino}-N-piperidin-4-yl-1H-pyrazole-3-carboxamide | | Authors: | Wyatt, P.G, Woodhead, A.J, Boulstridge, J.A, Berdini, V, Carr, M.G, Cross, D.M, Danillon, D, Davis, D.J, Devine, L.A, Early, T.R, Feltell, R.E, Lewis, E.J, McMenamin, R.L, Navarro, E.F, O'Brien, M.A, O'Reilly, M, Reule, M, Saxty, G, Seavers, L.C.A, Smith, D, Squires, M.S, Trewartha, G, Walker, M.T, Woolford, A.J. | | Deposit date: | 2008-05-15 | | Release date: | 2008-08-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of N-(4-Piperidinyl)-4-(2,6-Dichlorobenzoylamino)-1H-Pyrazole-3-Carboxamide (at7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design.
J.Med.Chem., 51, 2008
|
|
2W1H
 
 | | Fragment-Based Discovery of the Pyrazol-4-yl urea (AT9283), a Multi- targeted Kinase Inhibitor with Potent Aurora Kinase Activity | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, N-[3-(1H-BENZIMIDAZOL-2-YL)-1H-PYRAZOL-4-YL]BENZAMIDE | | Authors: | Howard, S, Berdini, V, Boulstridge, J.A, Carr, M.G, Cross, D.M, Curry, J, Devine, L.A, Early, T.R, Fazal, L, Gill, A.L, Heathcote, M, Maman, S, Matthews, J.E, McMenamin, R.L, Navarro, E.F, O'Brien, M.A, O'Reilly, M, Rees, D.C, Reule, M, Tisi, D, Williams, G, Vinkovic, M, Wyatt, P.G. | | Deposit date: | 2008-10-17 | | Release date: | 2009-01-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Fragment-Based Discovery of the Pyrazol-4-Yl Urea (at9283), a Multitargeted Kinase Inhibitor with Potent Aurora Kinase Activity.
J.Med.Chem., 52, 2009
|
|
2VTA
 
 | | Identification of N-(4-piperidinyl)-4-(2,6-dichlorobenzoylamino)-1H- pyrazole-3-carboxamide (AT7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design. | | Descriptor: | 1H-indazole, CELL DIVISION PROTEIN KINASE 2, GLYCEROL | | Authors: | Wyatt, P.G, Woodhead, A.J, Boulstridge, J.A, Berdini, V, Carr, M.G, Cross, D.M, Danillon, D, Davis, D.J, Devine, L.A, Early, T.R, Feltell, R.E, Lewis, E.J, McMenamin, R.L, Navarro, E.F, O'Brien, M.A, O'Reilly, M, Reule, M, Saxty, G, Seavers, L.C.A, Smith, D, Squires, M.S, Trewartha, G, Walker, M.T, Woolford, A.J. | | Deposit date: | 2008-05-13 | | Release date: | 2008-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of N-(4-Piperidinyl)-4-(2,6-Dichlorobenzoylamino)-1H-Pyrazole-3-Carboxamide (at7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design.
J.Med.Chem., 51, 2008
|
|
2WKA
 
 | | Structure of Plp_Thr_decanoyl-CoA aldimine form of Vibrio cholerae CqsA | | Descriptor: | CAI-1 AUTOINDUCER SYNTHASE, CHLORIDE ION, SULFATE ION, ... | | Authors: | Jahan, N, Potter, J.A, Sheikh, M.A, Botting, C.H, Shirran, S.L, Westwood, N.J, Taylor, G.L. | | Deposit date: | 2009-06-08 | | Release date: | 2009-07-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Insights Into the Biosynthesis of the Vibrio Cholerae Major Autoinducer Cai-1 from the Crystal Structure of the Plp-Dependent Enzyme Cqsa.
J.Mol.Biol., 392, 2009
|
|
1PIW
 
 | | APO AND HOLO STRUCTURES OF AN NADP(H)-DEPENDENT CINNAMYL ALCOHOL DEHYDROGENASE FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | Hypothetical zinc-type alcohol dehydrogenase-like protein in PRE5-FET4 intergenic region, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ZINC ION | | Authors: | Valencia, E, Larroy, C, Ochoa, W.F, Pares, X, Fita, I, Biosca, J.A. | | Deposit date: | 2003-05-30 | | Release date: | 2004-08-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Apo and Holo Structures of an NADP(H)-dependent Cinnamyl Alcohol Dehydrogenase from Saccharomyces cerevisiae
J.Mol.Biol., 341, 2004
|
|
2WWG
 
 | | Plasmodium falciparum thymidylate kinase in complex with dGMP and ADP | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Whittingham, J.L, Carrero-Lerida, J, Brannigan, J.A, Ruiz-Perez, L.M, Silva, A.P, Fogg, M.J, Wilkinson, A.J, Gilbert, I.H, Wilson, K.S, Gonzalez-Pacanowska, D. | | Deposit date: | 2009-10-23 | | Release date: | 2010-04-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for the Efficient Phosphorylation of Aztmp and Dgmp by Plasmodium Falciparum Type I Thymidylate Kinase.
Biochem.J., 428, 2010
|
|
1Q80
 
 | | Solution structure and dynamics of Nereis sarcoplasmic calcium binding protein | | Descriptor: | Sarcoplasmic calcium-binding protein | | Authors: | Rabah, G, Popescu, R, Cox, J.A, Engelborghs, Y, Craescu, C.T. | | Deposit date: | 2003-08-20 | | Release date: | 2004-09-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and internal dynamics of NSCP, a compact calcium-binding protein.
Febs J., 272, 2005
|
|
2WMA
 
 | |
2YOJ
 
 | | HCV NS5B polymerase complexed with pyridonylindole compound | | Descriptor: | 4-fluoranyl-6-[(7-fluoranyl-4-oxidanylidene-3H-quinazolin-6-yl)methyl]-8-(2-oxidanylidene-1H-pyridin-3-yl)furo[2,3-e]indole-7-carboxylic acid, PHOSPHATE ION, RNA-DIRECTED RNA POLYMERASE | | Authors: | Chen, K.X, Venkatraman, S, Anilkumar, G.N, Zeng, Q, Lesburg, C.A, Vibulbhan, B, Yang, W, Velazquez, F, Chan, T.-Y, Bennett, F, Sannigrahi, M, Jiang, Y, Duca, J.S, Pinto, P, Gavalas, S, Huang, Y, Wu, W, Selyutin, O, Agrawal, S, Feld, B, Huang, H.-C, Li, C, Cheng, K.-C, Shih, N.-Y, Kozlowski, J.A, Rosenblum, S.B, Njoroge, F.G. | | Deposit date: | 2012-10-24 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Discovery of Sch 900188: A Potent Hepatitis C Virus Ns5B Polymerase Inhibitor Prodrug as a Development Candidate
Acs Med.Chem.Lett., 5, 2014
|
|
2YDK
 
 | | Discovery of Checkpoint Kinase Inhibitor AZD7762 by Structure Based Design and Optimization of Thiophene Carboxamide Ureas | | Descriptor: | 2-(CARBAMOYLAMINO)-5-PHENYL-N-[(3S)-PIPERIDIN-3-YL]THIOPHENE-3-CARBOXAMIDE, GLYCEROL, SERINE/THREONINE-PROTEIN KINASE CHK1, ... | | Authors: | Read, J.A, Breed, J, Haye, H, McCall, E, Rowsell, S, Vallentine, A, White, A. | | Deposit date: | 2011-03-22 | | Release date: | 2012-04-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Checkpoint Kinase Inhibitor (S)-5-(3-Fluorophenyl)-N-(Piperidin-3-Yl)-3-Ureidothiophene-2-Carboxamide (Azd7762) by Structure-Based Design and Optimization of Thiophenecarboxamide Ureas.
J.Med.Chem., 55, 2012
|
|
1TYF
 
 | |
4APE
 
 | | THE ACTIVE SITE OF ASPARTIC PROTEINASES | | Descriptor: | ENDOTHIAPEPSIN | | Authors: | Pearl, L.H, Sewell, B.T, Jenkins, J.A, Cooper, J.B, Blundell, T.L. | | Deposit date: | 1986-06-09 | | Release date: | 1986-07-14 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Active Site of Aspartic Proteinases
FEBS Lett., 174, 1984
|
|
2WCL
 
 | | Structure of BMori GOBP2 (General Odorant Binding Protein 2) with (8E, 10Z)-hexadecadien-1-ol | | Descriptor: | (8E,10Z)-HEXADECA-8,10-DIEN-1-OL, GENERAL ODORANT-BINDING PROTEIN 1, MAGNESIUM ION | | Authors: | Robertson, G, Zhou, J.-J, He, X, Pickett, J.A, Field, L.M, Keep, N.H. | | Deposit date: | 2009-03-12 | | Release date: | 2009-08-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Characterisation of Bombyx Mori Odorant-Binding Proteins Reveals that a General Odorant-Binding Protein Discriminates between Sex Pheromone Components.
J.Mol.Biol., 389, 2009
|
|
2WAJ
 
 | | Crystal structure of human Jnk3 complexed with a 1-aryl-3,4- dihydroisoquinoline inhibitor | | Descriptor: | 1-(3-BROMOPHENYL)-7-CHLORO-6-METHOXY-3,4-DIHYDROISOQUINOLINE, MITOGEN-ACTIVATED PROTEIN KINASE 10 | | Authors: | Bax, B.D, Christopher, J.A, Jones, E.J, Mosley, J.E. | | Deposit date: | 2009-02-08 | | Release date: | 2009-03-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 1-Aryl-3,4-Dihydroisoquinoline Inhibitors of Jnk3.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
2VZL
 
 | | FERREDOXIN-NADP REDUCTASE (MUTATIONS: T155G, A160T, L263P AND Y303S) COMPLEXED WITH NAD BY COCRYSTALLIZATION | | Descriptor: | FERREDOXIN-NADP REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Herguedas, B, Martinez-Julvez, M, Hermoso, J.A, Peregrina, J.R, Medina, M. | | Deposit date: | 2008-07-28 | | Release date: | 2009-04-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Protein Motifs Involved in Coenzyme Interaction and Enzymatic Efficiency in Anabaena Ferredoxin-Nadp+ Reductase.
Biochemistry, 48, 2009
|
|
1UJL
 
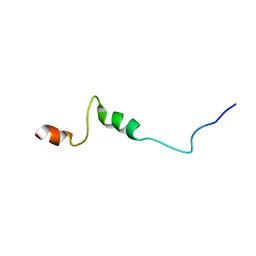 | | Solution Structure of the HERG K+ channel S5-P extracellular linker | | Descriptor: | Potassium voltage-gated channel subfamily H member 2 | | Authors: | Torres, A.M, Bansal, P.S, Sunde, M, Clarke, C.E, Bursill, J.A, Smith, D.J, Bauskin, A, Breit, S.N, Campbell, T.J, Alewood, P.F, Kuchel, P.W, Vandenberg, J.I. | | Deposit date: | 2003-08-05 | | Release date: | 2003-11-04 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of the HERG K+ channel S5P extracellular linker: role of an amphipathic alpha-helix
in C-type inactivation.
J.Biol.Chem., 278, 2003
|
|
2WDU
 
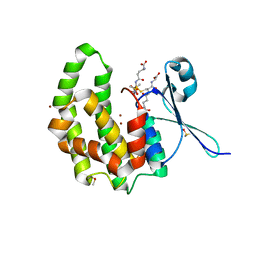 | | Fasciola hepatica sigma class GST | | Descriptor: | BROMIDE ION, DIMETHYL SULFOXIDE, GLUTATHIONE, ... | | Authors: | Line, K, Isupov, M.N, LaCourse, E.J, Brophy, P.M, Littlechild, J.A. | | Deposit date: | 2009-03-26 | | Release date: | 2010-04-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | The 1.6 Angstrom Crystal Structure of the Fasciola Hepatica Sigma Class Gst
To be Published
|
|
2WC6
 
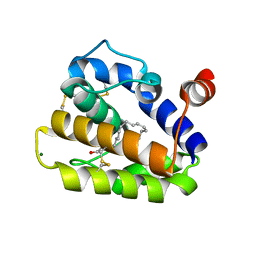 | | Structure of BMori GOBP2 (General Odorant Binding Protein 2) with bombykol and water to Arg 110 | | Descriptor: | GENERAL ODORANT-BINDING PROTEIN 1, HEXADECA-10,12-DIEN-1-OL, MAGNESIUM ION | | Authors: | Robertson, G, Zhou, J.-J, He, X, Pickett, J.A, Field, L.M, Keep, N.H. | | Deposit date: | 2009-03-09 | | Release date: | 2009-08-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterisation of Bombyx Mori Odorant-Binding Proteins Reveals that a General Odorant-Binding Protein Discriminates between Sex Pheromone Components.
J.Mol.Biol., 389, 2009
|
|
2W54
 
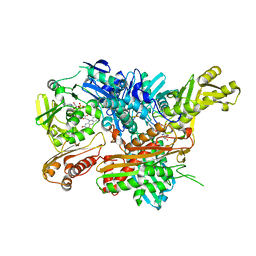 | | Crystal Structure of Xanthine Dehydrogenase from Rhodobacter capsulatus in Complex with Bound Inhibitor Pterin-6-aldehyde | | Descriptor: | 6-HYDROXYMETHYLPTERIN, BARIUM ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Doebbler, J.A, Truglio, J.J, Leimkuhler, S, Kisker, C. | | Deposit date: | 2008-12-04 | | Release date: | 2008-12-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Mechanism of Substrate and Inhibitor Binding of Rhodobacter Capsulatus Xanthine Dehydrogenase.
J.Biol.Chem., 284, 2009
|
|
2WB9
 
 | | Fasciola hepatica sigma class GST | | Descriptor: | BROMIDE ION, CYSTEINE, GLUTATHIONE, ... | | Authors: | Line, K, Isupov, M.N, LaCourse, J, Brophy, P.M, Littlechild, J.A. | | Deposit date: | 2009-02-23 | | Release date: | 2010-03-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | The 1.6 Angstrom Crystal Structure of the Fasciola Hepatica Sigma Class Gst
To be Published
|
|
1UUG
 
 | | ESCHERICHIA COLI URACIL-DNA GLYCOSYLASE:INHIBITOR COMPLEX WITH WILD-TYPE UDG AND WILD-TYPE UGI | | Descriptor: | URACIL-DNA GLYCOSYLASE, URACIL-DNA GLYCOSYLASE INHIBITOR | | Authors: | Mol, C.D, Arvai, A.S, Putnam, C.D, Tainer, J.A. | | Deposit date: | 1998-10-31 | | Release date: | 1999-03-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Protein mimicry of DNA from crystal structures of the uracil-DNA glycosylase inhibitor protein and its complex with Escherichia coli uracil-DNA glycosylase
J.Mol.Biol., 287, 1999
|
|
1W3F
 
 | | Crystal structure of the hemolytic lectin from the mushroom Laetiporus sulphureus complexed with N-acetyllactosamine in the gamma motif | | Descriptor: | GLYCEROL, HEMOLYTIC LECTIN FROM LAETIPORUS SULPHUREUS, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Mancheno, J.M, Tateno, H, Goldstein, I.J, Martinez-Ripoll, M, Hermoso, J.A. | | Deposit date: | 2004-07-15 | | Release date: | 2005-02-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural Analysis of the Laetiporus Sulphureus Hemolytic Pore-Forming Lectin in Complex with Sugars
J.Biol.Chem., 280, 2005
|
|
2X5N
 
 | | Crystal Structure of the SpRpn10 VWA domain | | Descriptor: | 26S PROTEASOME REGULATORY SUBUNIT RPN10, SULFATE ION | | Authors: | Riedinger, C, Boehringer, J, Trempe, J.-F, Lowe, E.D, Brown, N.R, Gehring, K, Noble, M.E.M, Gordon, C, Endicott, J.A. | | Deposit date: | 2010-02-10 | | Release date: | 2010-08-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The Structure of Rpn10 and its Interactions with Polyubiquitin Chains and the Proteasome Subunit Rpn12.
J.Biol.Chem., 285, 2010
|
|
