4RLP
 
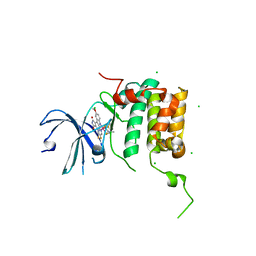 | | Human p70s6k1 with ruthenium-based inhibitor FL772 | | Descriptor: | CHLORIDE ION, [(amino-kappaN)methanethiolato](3-fluoro-9-methoxypyrido[2,3-a]pyrrolo[3,4-c]carbazole-5,7(6H,12H)-dionato-kappa~2~N,N')(N-methyl-1,4,7-trithiecan-9-amine-kappa~3~S~1~,S~4~,S~7~)ruthenium, p70S6K1 | | Authors: | Domsic, J.F, Barber-Rotenberg, J, Salami, J, Qin, J, Marmorstein, R. | | Deposit date: | 2014-10-17 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Development of Organometallic S6K1 Inhibitors.
J.Med.Chem., 58, 2015
|
|
7N84
 
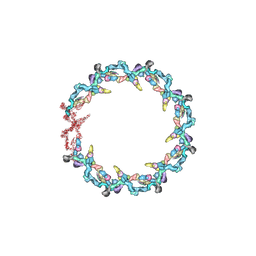 | | Double nuclear outer ring from the isolated yeast NPC | | Descriptor: | Nucleoporin 145c, Nucleoporin NUP120, Nucleoporin NUP133, ... | | Authors: | Akey, C.W, Rout, M.P, Ouch, C, Echevarria, I, Fernandez-Martinez, J, Nudelman, I. | | Deposit date: | 2021-06-13 | | Release date: | 2022-01-26 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (11.6 Å) | | Cite: | Comprehensive structure and functional adaptations of the yeast nuclear pore complex.
Cell, 185, 2022
|
|
8ZHE
 
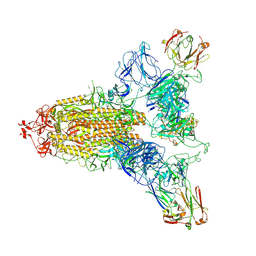 | | SARS-CoV-2 spike trimer (6P) in complex with three R1-26 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-26 Fab, ... | | Authors: | Yan, Q, Gao, X, Liu, B, Hou, R, He, P, Li, Z, Chen, Q, Wang, J, He, J, Chen, L, Zhao, J, Xiong, X. | | Deposit date: | 2024-05-10 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Antibodies utilizing VL6-57 light chains target a convergent cryptic epitope on SARS-CoV-2 spike protein and potentially drive the genesis of Omicron variants.
Nat Commun, 15, 2024
|
|
8ZHP
 
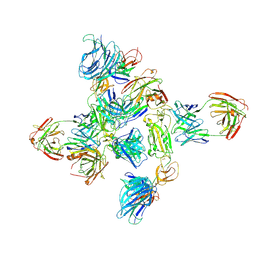 | | Dimer of SARS-CoV-2 S1 in complex with H18 and R1-32 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of H18 Fab, Heavy chain of R1-32 Fab, ... | | Authors: | Yan, Q, Gao, X, Liu, B, Hou, R, He, P, Li, Z, Chen, Q, Wang, J, He, J, Chen, L, Zhao, J, Xiong, X. | | Deposit date: | 2024-05-11 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Antibodies utilizing VL6-57 light chains target a convergent cryptic epitope on SARS-CoV-2 spike protein and potentially drive the genesis of Omicron variants.
Nat Commun, 15, 2024
|
|
8ZHF
 
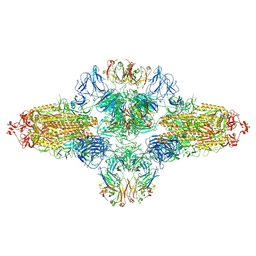 | | SARS-CoV-2 spike trimer (6P) in complex with R1-26 Fab, head-to-head aggregate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-26 Fab, ... | | Authors: | Yan, Q, Gao, X, Liu, B, Hou, R, He, P, Li, Z, Chen, Q, Wang, J, He, J, Chen, L, Zhao, J, Xiong, X. | | Deposit date: | 2024-05-10 | | Release date: | 2024-08-21 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (5.26 Å) | | Cite: | Antibodies utilizing VL6-57 light chains target a convergent cryptic epitope on SARS-CoV-2 spike protein and potentially drive the genesis of Omicron variants.
Nat Commun, 15, 2024
|
|
6ROY
 
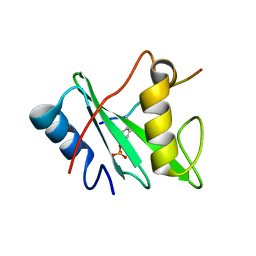 | |
5IZ6
 
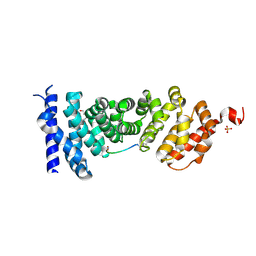 | | Protein-protein interaction | | Descriptor: | Adenomatous polyposis coli protein, DI(HYDROXYETHYL)ETHER, PHQ-ALA-GLY-GLU-ALA-LEU-TYR-GLU-NH2, ... | | Authors: | Zhao, Y, Jiang, H, Yang, X, Jiang, F, Song, K, Zhang, J. | | Deposit date: | 2016-03-25 | | Release date: | 2017-07-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Peptidomimetic inhibitors of APC-Asef interaction block colorectal cancer migration.
Nat. Chem. Biol., 13, 2017
|
|
7UFJ
 
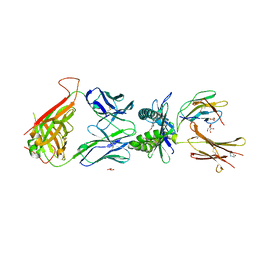 | | Structure of human MR1-ethylvanillin in complex with human MAIT A-F7 TCR | | Descriptor: | 3-ethoxy-4-hydroxybenzaldehyde, ACETATE ION, Beta-2-microglobulin, ... | | Authors: | Wang, C.J, Rossjohn, J, Le Nours, J. | | Deposit date: | 2022-03-22 | | Release date: | 2022-12-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Quantitative affinity measurement of small molecule ligand binding to Major Histocompatibility Complex class-I related protein 1 MR1.
J.Biol.Chem., 298, 2022
|
|
4RLO
 
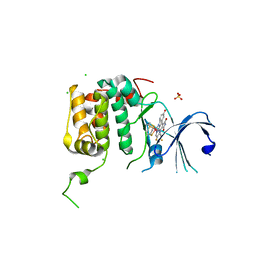 | | Human p70s6k1 with ruthenium-based inhibitor EM5 | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Domsic, J.F, Barber-Rotenberg, J, Salami, J, Qin, J, Marmorstein, R. | | Deposit date: | 2014-10-17 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.527 Å) | | Cite: | Development of Organometallic S6K1 Inhibitors.
J.Med.Chem., 58, 2015
|
|
5LSP
 
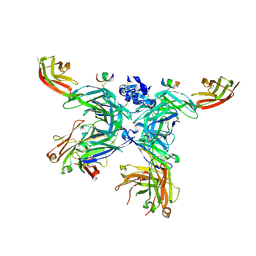 | | 107_A07 Fab in complex with fragment of the Met receptor | | Descriptor: | 107_A07 Fab heavy chain, 107_A07 Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | DiCara, D, Chirgadze, D.Y, Pope, A, Karatt-Vellatt, A, Winter, A, van den Heuvel, J, Gherardi, E, McCafferty, J. | | Deposit date: | 2016-09-05 | | Release date: | 2017-09-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.605 Å) | | Cite: | Characterization and structural determination of a new anti-MET function-blocking antibody with binding epitope distinct from the ligand binding domain.
Sci Rep, 7, 2017
|
|
8I5M
 
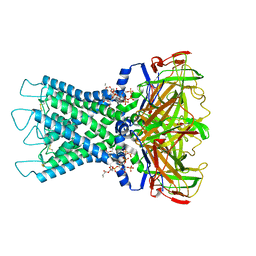 | | Rat Kir4.1 in complex with PIP2 | | Descriptor: | ATP-sensitive inward rectifier potassium channel 10, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Zhao, C, Guo, J. | | Deposit date: | 2023-01-26 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Pharmacological inhibition of Kir4.1 evokes rapid-onset antidepressant responses.
Nat.Chem.Biol., 20, 2024
|
|
3B3O
 
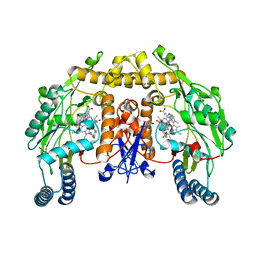 | | Structure of neuronal nos heme domain in complex with a inhibitor (+-)-n1-{cis-4'-[(6"-amino-4"-methylpyridin-2"-yl)methyl]pyrrolidin-3'-yl}-n2-(4'-chlorobenzyl)ethane-1,2-diamine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N-{(3S,4S)-4-[(6-AMINO-4-METHYLPYRIDIN-2-YL)METHYL]PYRROLIDIN-3-YL}-N'-(4-CHLOROBENZYL)ETHANE-1,2-DIAMINE, ... | | Authors: | Igarashi, J, Li, H, Poulos, T.L. | | Deposit date: | 2007-10-22 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of constitutive nitric oxide synthases in complex with de novo designed inhibitors.
J.Med.Chem., 52, 2009
|
|
8I5N
 
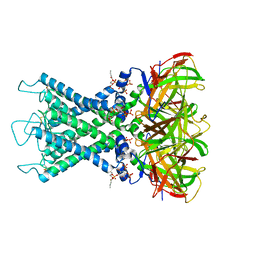 | | Rat Kir4.1 in complex with PIP2 and Lys05 | | Descriptor: | ATP-sensitive inward rectifier potassium channel 10, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Zhao, C, Guo, J. | | Deposit date: | 2023-01-26 | | Release date: | 2024-02-28 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Pharmacological inhibition of Kir4.1 evokes rapid-onset antidepressant responses.
Nat.Chem.Biol., 20, 2024
|
|
8B22
 
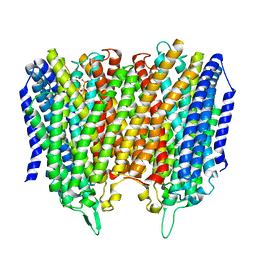 | |
8B23
 
 | |
8B21
 
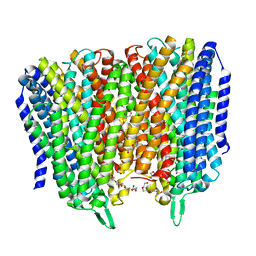 | | Time-resolved structure of K+-dependent Na+-PPase from Thermotoga maritima 0-60-seconds post reaction initiation with Na+ | | Descriptor: | DI(HYDROXYETHYL)ETHER, DODECYL-BETA-D-MALTOSIDE, K(+)-stimulated pyrophosphate-energized sodium pump, ... | | Authors: | Strauss, J, Vidilaseris, K, Goldman, A. | | Deposit date: | 2022-09-12 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Functional and structural asymmetry suggest a unifying principle for catalysis in membrane-bound pyrophosphatases.
Embo Rep., 25, 2024
|
|
8B24
 
 | | Time-resolved structure of K+-dependent Na+-PPase from Thermotoga maritima 3600-seconds post reaction initiation with Na+ | | Descriptor: | DIPHOSPHATE, K(+)-stimulated pyrophosphate-energized sodium pump, MAGNESIUM ION, ... | | Authors: | Strauss, J, Vidilaseris, K, Goldman, A. | | Deposit date: | 2022-09-12 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (4.53 Å) | | Cite: | Functional and structural asymmetry suggest a unifying principle for catalysis in membrane-bound pyrophosphatases.
Embo Rep., 25, 2024
|
|
8IGA
 
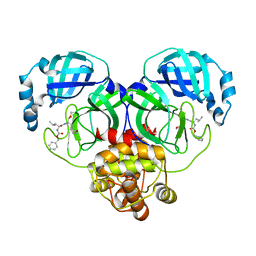 | |
5IW5
 
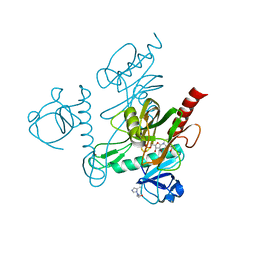 | |
6SKK
 
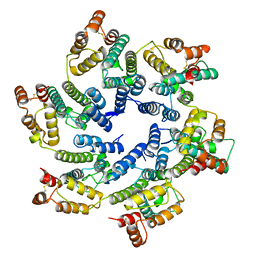 | | Structure of the native full-length HIV-1 capsid protein in helical assembly (-13,8) | | Descriptor: | capsid protein | | Authors: | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | Deposit date: | 2019-08-15 | | Release date: | 2020-08-26 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8XM3
 
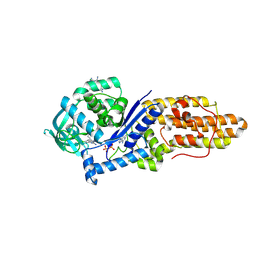 | | Methionyl-tRNA synthetase from Staphylococcus aureus complexed with a chlorhexidine derivative and ATP | | Descriptor: | 1-(4-chlorophenyl)-3-[~{N}-[4-[[~{N}-[~{N}-(4-chlorophenyl)carbamimidoyl]carbamimidoyl]amino]butyl]carbamimidoyl]guanidine, ADENOSINE-5'-TRIPHOSPHATE, Methionine--tRNA ligase | | Authors: | Lu, F, Xia, K, Yi, J, Chen, B, Luo, Z, Xu, J, Gu, Q, Zhou, H. | | Deposit date: | 2023-12-27 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Biochemical and structural characterization of chlorhexidine as an ATP-assisted inhibitor against type 1 methionyl-tRNA synthetase from Gram-positive bacteria.
Eur.J.Med.Chem., 268, 2024
|
|
7GMW
 
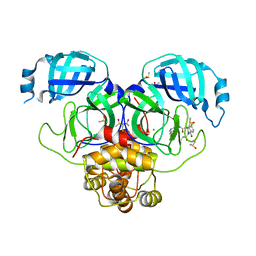 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-e119ab4f-1 (Mpro-P2224) | | Descriptor: | (4S)-6-chloro-N-(7-fluoroisoquinolin-4-yl)-2-[2-(methylamino)-2-oxoethyl]-1-oxo-1,2,3,4-tetrahydroisoquinoline-4-carboxamide, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.826 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GNJ
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with EDJ-MED-976a33d5-1 (Mpro-P2724) | | Descriptor: | 1-[(3'S)-6-chloro-1'-(isoquinolin-4-yl)-1,2'-dioxo-1H-spiro[isoquinoline-4,3'-pyrrolidin]-2(3H)-yl]-N-methylcyclopropane-1-carboxamide, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GLO
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with JAN-GHE-5a013bed-2 (Mpro-P2001) | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.927 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GM4
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with ALP-POS-6f6ae286-5 (Mpro-P2080) | | Descriptor: | (4S)-6-chloro-N-(7-fluoroisoquinolin-4-yl)-2-[2-(methylamino)-2-oxoethyl]-1,2,3,4-tetrahydroisoquinoline-4-carboxamide, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
