7O7E
 
 | | Crystal structure of rsEGFP2 mutant V151L in the fluorescent on-state determined by synchrotron radiation at 100K | | Descriptor: | Green fluorescent protein | | Authors: | Woodhouse, J, Adam, V, Hadjidemetriou, K, Colletier, J.-P, Weik, M. | | Deposit date: | 2021-04-13 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rational Control of Off-State Heterogeneity in a Photoswitchable Fluorescent Protein Provides Switching Contrast Enhancement.
Chemphyschem, 23, 2022
|
|
7O7D
 
 | | Crystal structure of rsEGFP2 mutant V151A in the fluorescent on-state determined by synchrotron radiation at 100K | | Descriptor: | Green fluorescent protein | | Authors: | Woodhouse, J, Adam, V, Hadjidemetriou, K, Colletier, J.-P, Weik, M. | | Deposit date: | 2021-04-13 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Rational Control of Off-State Heterogeneity in a Photoswitchable Fluorescent Protein Provides Switching Contrast Enhancement.
Chemphyschem, 23, 2022
|
|
3JZQ
 
 | |
7O7U
 
 | | Crystal structure of rsEGFP2 in the non-fluorescent off-state determined by serial femtosecond crystallography at room temperature | | Descriptor: | Green fluorescent protein | | Authors: | Hadjidemetriou, K, Woodhouse, J, Coquelle, N, Barends, T.R.M, Schlichting, I, Weik, M, Colletier, J.-P. | | Deposit date: | 2021-04-13 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Rational Control of Off-State Heterogeneity in a Photoswitchable Fluorescent Protein Provides Switching Contrast Enhancement.
Chemphyschem, 23, 2022
|
|
3ZD0
 
 | | The Solution Structure of Monomeric Hepatitis C Virus p7 Yields Potent Inhibitors of Virion Release | | Descriptor: | P7 PROTEIN | | Authors: | Foster, T.L, Sthompson, G, Kalverda, A.P, Kankanala, J, Thompson, J, Barker, A.M, Clarke, D, Noerenberg, M, Pearson, A.R, Rowlands, D.J, Homans, S.W, Harris, M, Foster, R, Griffin, S.D.C. | | Deposit date: | 2012-11-23 | | Release date: | 2013-09-04 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure-Guided Design Affirms Inhibitors of Hepatitis C Virus P7 as a Viable Class of Antivirals Targeting Virion Release
Hepatology, 59, 2014
|
|
7O7H
 
 | | Crystal structure of rsEGFP2 mutant V151L in the non-fluorescent off-state determined by synchrotron radiation at 100K | | Descriptor: | Green fluorescent protein | | Authors: | Woodhouse, J, Adam, V, Hadjidemetriou, K, Colletier, J.P, Weik, M. | | Deposit date: | 2021-04-13 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Rational Control of Off-State Heterogeneity in a Photoswitchable Fluorescent Protein Provides Switching Contrast Enhancement.
Chemphyschem, 23, 2022
|
|
7O7C
 
 | | Crystal structure of rsEGFP2 mutant V151A in the non-fluorescent off-state determined by synchrotron radiation at 100K | | Descriptor: | Green fluorescent protein, SULFATE ION | | Authors: | Woodhouse, J, Adam, V, Hadjidemetriou, K, Colletier, J.-P, Weik, M. | | Deposit date: | 2021-04-13 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Rational Control of Off-State Heterogeneity in a Photoswitchable Fluorescent Protein Provides Switching Contrast Enhancement.
Chemphyschem, 23, 2022
|
|
7O5R
 
 | | Crystal structure of holo-SwHPA-Mn (hydroxyketoacid aldolase) from Sphingomonas wittichii RW1 | | Descriptor: | BROMIDE ION, MANGANESE (II) ION, POTASSIUM ION, ... | | Authors: | Laustsen, J, Justo, I, Marsden, S.R, Hanefeld, U, Bento, I. | | Deposit date: | 2021-04-09 | | Release date: | 2022-10-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Substrate Induced Movement of the Metal Cofactor between Active and Resting State.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7O9R
 
 | | Crystal structure of holo-H44A mutant of Hydroxy ketone aldolase (SwHKA) from Sphingomonas wittichii RW1 | | Descriptor: | BROMIDE ION, DI(HYDROXYETHYL)ETHER, HpcH/HpaI aldolase, ... | | Authors: | Laustsen, J, Justo, I, Marsden, S.R, Hanefeld, U, Bento, I. | | Deposit date: | 2021-04-16 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate Induced Movement of the Metal Cofactor between Active and Resting State.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
4EQ8
 
 | | Crystal structure of PA1844 from Pseudomonas aeruginosa PAO1 | | Descriptor: | GLYCEROL, Putative uncharacterized protein | | Authors: | Shang, G, Li, N, Zhang, J, Lu, D, Yu, Q, Zhao, Y, Liu, X, Xu, S, Gu, L. | | Deposit date: | 2012-04-18 | | Release date: | 2012-09-12 | | Last modified: | 2013-07-24 | | Method: | X-RAY DIFFRACTION (1.392 Å) | | Cite: | Structural insight into how Pseudomonas aeruginosa peptidoglycanhydrolase Tse1 and its immunity protein Tsi1 function.
Biochem.J., 448, 2012
|
|
3DX7
 
 | | Crystal Structure of HLA-B*4403 presenting 10mer EBV antigen | | Descriptor: | ACETATE ION, Beta-2-microglobulin, EBV decapeptide epitope, ... | | Authors: | Archbold, J.K, Ely, L.K, Rossjohn, J. | | Deposit date: | 2008-07-23 | | Release date: | 2009-01-27 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Natural micropolymorphism in human leukocyte antigens provides a basis for genetic control of antigen recognition.
J.Exp.Med., 206, 2009
|
|
7O87
 
 | | Crystal structure of holo-F210W mutant of Hydroxy ketone aldolase (SwHKA) from Sphingomonas wittichii RW1 in complex with hydroxypyruvate | | Descriptor: | 3-HYDROXYPYRUVIC ACID, BROMIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Laustsen, J, Justo, I, Marsden, S.R, Hanefeld, U, Bento, I. | | Deposit date: | 2021-04-14 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Substrate Induced Movement of the Metal Cofactor between Active and Resting State.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7O5W
 
 | | Crystal structure of holo-F210W mutant of Hydroxy ketone aldolase (SwHKA)from Sphingomonas wittichii RW1 | | Descriptor: | BROMIDE ION, DI(HYDROXYETHYL)ETHER, HpcH/HpaI aldolase, ... | | Authors: | Laustsen, J, Justo, I, Marsden, S.R, Hanefeld, U, Bento, I. | | Deposit date: | 2021-04-09 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Substrate Induced Movement of the Metal Cofactor between Active and Resting State.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7OCJ
 
 | | Crystal structure of E.coli LexA in complex with nanobody NbSOS2(Nb14509) | | Descriptor: | 1,2-ETHANEDIOL, LexA repressor, NbSOS2 (14509) | | Authors: | Maso, L, Vascon, F, Chinellato, M, Pardon, E, Steyaert, J, Angelini, A, Tondi, D, Cendron, L. | | Deposit date: | 2021-04-27 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Nanobodies targeting LexA autocleavage disclose a novel suppression strategy of SOS-response pathway.
Structure, 30, 2022
|
|
1GIC
 
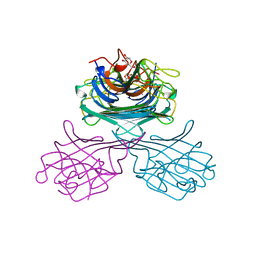 | | CONCANAVALIN A COMPLEXED WITH METHYL ALPHA-D-GLUCOPYRANOSIDE | | Descriptor: | CALCIUM ION, CONCANAVALIN A, MANGANESE (II) ION, ... | | Authors: | Bradbrook, G.M, Gleichmann, T, Harrop, S.J, Helliwell, J.R, Habash, J, Kalb(Gilboa), A.J, Tong, L, Wan, T.C.M, Yariv, J. | | Deposit date: | 1996-08-08 | | Release date: | 1997-08-20 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure solution of a cubic crystal of concanavalin A complexed with methyl alpha-D-glucopyranoside.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
7OBU
 
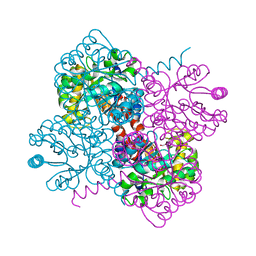 | | Crystal structure of holo-F210W mutant of Hydroxy ketone aldolase (SwHKA) from Sphingomonas wittichii RW1, with the active site in the resting and the active state | | Descriptor: | 3-HYDROXYPYRUVIC ACID, HpcH/HpaI aldolase, MAGNESIUM ION, ... | | Authors: | Laustsen, J, Justo, I, Marsden, S.R, Hanefeld, U, Bento, I. | | Deposit date: | 2021-04-23 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Substrate Induced Movement of the Metal Cofactor between Active and Resting State.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
3O8V
 
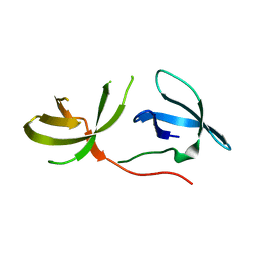 | | Crystal Structure of the Tudor Domains from FXR1 | | Descriptor: | Fragile X mental retardation syndrome-related protein 1 | | Authors: | Lam, R, Guo, Y.H, Adams-Cioaba, M, Bian, C.B, Mackenzie, F, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-08-03 | | Release date: | 2010-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Studies of the Tandem Tudor Domains of Fragile X Mental Retardation Related Proteins FXR1 and FXR2.
Plos One, 5, 2010
|
|
3ZDT
 
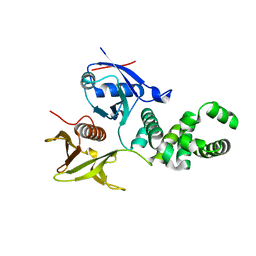 | | Crystal structure of basic patch mutant FAK FERM domain FAK31- 405 K216A, K218A, R221A, K222A | | Descriptor: | FOCAL ADHESION KINASE 1 | | Authors: | Goni, G.M, Epifano, C, Boskovic, J, Camacho-Artacho, M, Zhou, J, Martin, M.T, Eck, M.J, Kremer, L, Graeter, F, Gervasio, F.L, Perez-Moreno, M, Lietha, D. | | Deposit date: | 2012-11-30 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Phosphatidylinositol 4,5-Bisphosphate Triggers Activation of Focal Adhesion Kinase by Inducing Clustering and Conformational Changes.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7OA4
 
 | | Crystal structure of the N-terminal endonuclease domain of La Crosse virus L-protein bound to compound L-742,001 | | Descriptor: | (2Z)-4-[1-benzyl-4-(4-chlorobenzyl)piperidin-4-yl]-2-hydroxy-4-oxobut-2-enoic acid, FORMIC ACID, MANGANESE (II) ION, ... | | Authors: | Feracci, M, Hernandez, S, Vincentelli, R, Ferron, F, Reguera, J, Canard, B, Alvarez, K. | | Deposit date: | 2021-04-19 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the N-terminal endonuclease domain of La Crosse virus L-protein bound to compound L-742,001
To Be Published
|
|
2P8E
 
 | | Crystal structure of the serine/threonine phosphatase domain of human PPM1B | | Descriptor: | MAGNESIUM ION, PPM1B beta isoform variant 6 | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Lau, C, Xu, W, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-22 | | Release date: | 2007-04-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.816 Å) | | Cite: | Structural genomics of protein phosphatases.
J.Struct.Funct.Genom., 8, 2007
|
|
1FYT
 
 | | CRYSTAL STRUCTURE OF A COMPLEX OF A HUMAN ALPHA/BETA-T CELL RECEPTOR, INFLUENZA HA ANTIGEN PEPTIDE, AND MHC CLASS II MOLECULE, HLA-DR1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HEMAGGLUTININ HA1 PEPTIDE CHAIN, HLA CLASS II HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Hennecke, J, Carfi, A, Wiley, D.C. | | Deposit date: | 2000-10-03 | | Release date: | 2000-11-08 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of a covalently stabilized complex of a human alphabeta T-cell receptor, influenza HA peptide and MHC class II molecule, HLA-DR1.
EMBO J., 19, 2000
|
|
4PJ5
 
 | | Structure of human MR1-Ac-6-FP in complex with human MAIT TRBV6-1 TCR | | Descriptor: | Beta-2-microglobulin, Major histocompatibility complex class I-related gene protein, N-(6-formyl-4-oxo-3,4-dihydropteridin-2-yl)acetamide, ... | | Authors: | Birkinshaw, R.W, Rossjohn, J. | | Deposit date: | 2014-05-12 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A molecular basis underpinning the T cell receptor heterogeneity of mucosal-associated invariant T cells.
J.Exp.Med., 211, 2014
|
|
6BBD
 
 | | Structure of N-glycosylated porcine surfactant protein-D complexed with glycerol | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | van Eijk, M, Rynkiewicz, M.J, Khatri, K, Leymarie, N, Zaia, J, White, M.R, Hartshorn, K.L, Cafarella, T.R, van Die, I, Hessing, M, Seaton, B.A, Haagsman, H.P. | | Deposit date: | 2017-10-18 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Lectin-mediated binding and sialoglycans of porcine surfactant protein D synergistically neutralize influenza A virus.
J. Biol. Chem., 293, 2018
|
|
2P65
 
 | | Crystal Structure of the first nucleotide binding domain of chaperone ClpB1, putative, (Pv089580) from Plasmodium Vivax | | Descriptor: | Hypothetical protein PF08_0063 | | Authors: | Wernimont, A.K, Lew, J, Kozieradzki, I, Lin, Y.H, Hassanali, A, Zhao, Y, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Artz, J.D, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-03-16 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the first nucleotide binding domain of chaperone ClpB1, putative, (Pv089580) from Plasmodium Vivax
To be Published
|
|
1SJJ
 
 | |
