3FK9
 
 | | Crystal structure of mMutator MutT protein from Bacillus halodurans | | Descriptor: | Mutator MutT protein | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Hu, S, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-12-16 | | Release date: | 2009-01-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of mMutator MutT protein from Bacillus halodurans
To be Published
|
|
1F4Q
 
 | | CRYSTAL STRUCTURE OF APO GRANCALCIN | | Descriptor: | GRANCALCIN | | Authors: | Jia, J, Han, Q, Borregaard, N, Lollike, K, Cygler, M. | | Deposit date: | 2000-06-08 | | Release date: | 2000-09-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human grancalcin, a member of the penta-EF-hand protein family.
J.Mol.Biol., 300, 2000
|
|
6ADX
 
 | | Crystal structure of the Zika virus NS3 helicase (ADP-Mn2+ complex, form 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MANGANESE (II) ION, Serine protease NS3 | | Authors: | Fang, J, Lu, G, Gong, P. | | Deposit date: | 2018-08-02 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Crystallographic Snapshots of the Zika Virus NS3 Helicase Help Visualize the Reactant Water Replenishment.
ACS Infect Dis, 5, 2019
|
|
3F2Z
 
 | | Crystal structure of the C-terminal domain of a chitobiase (BF3579) from Bacteroides fragilis, Northeast Structural Genomics Consortium Target BfR260B | | Descriptor: | uncharacterized protein BF3579 | | Authors: | Forouhar, F, Lew, S, Seetharaman, J, Janjua, H, Xiao, R, Foote, E.L, Ciccosanti, C, Lee, D, Nair, R, Everett, J.K, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-10-30 | | Release date: | 2008-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of the C-terminal domain of a chitobiase (BF3579) from Bacteroides fragilis, Northeast Structural Genomics Consortium Target BfR260B
To be Published
|
|
3FKU
 
 | | Crystal structure of influenza hemagglutinin (H5) in complex with a broadly neutralizing antibody F10 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, Neutralizing antibody F10, ... | | Authors: | Hwang, W.C, Santelli, E, Stec, B, Wei, G, Cadwell, G, Bankston, L.A, Sui, J, Perez, S, Aird, D, Chen, L.M, Ali, M, Murakami, A, Yammanuru, A, Han, T, Cox, N, Donis, R.O, Liddington, R.C, Marasco, W.A. | | Deposit date: | 2008-12-17 | | Release date: | 2009-02-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses.
Nat.Struct.Mol.Biol., 16, 2009
|
|
3F4W
 
 | |
6AGZ
 
 | | Crystal structure of Old Yellow Enzyme from Pichia sp. AKU4542 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Old Yellow Enzyme | | Authors: | Horita, S, Kataoka, M, Kitamura, N, Nakagawa, T, Miyakawa, T, Ohtsuka, J, Nagata, K, Shimizu, S, Tanokura, M. | | Deposit date: | 2018-08-15 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of different substrate preferences of two old yellow enzymes from yeasts in the asymmetric reduction of enone compounds.
Biosci.Biotechnol.Biochem., 83, 2019
|
|
6AHY
 
 | | Wnt signaling complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Hirai, H, Arimori, T, Matoba, K, Mihara, E, Takagi, J. | | Deposit date: | 2018-08-21 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a mammalian Wnt-frizzled complex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
3F6L
 
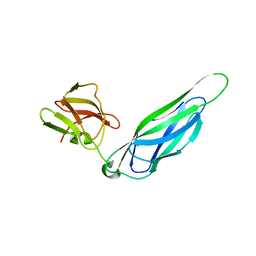 | | Structure of the F4 fimbrial chaperone FaeE | | Descriptor: | Chaperone protein faeE | | Authors: | Van Molle, I, Moonens, K, Buts, L, Garcia-Pino, A, Wyns, L, De Greve, H, Bouckaert, J. | | Deposit date: | 2008-11-06 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | The F4 fimbrial chaperone FaeE is stable as a monomer that does not require self-capping of its pilin-interactive surfaces
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3FOK
 
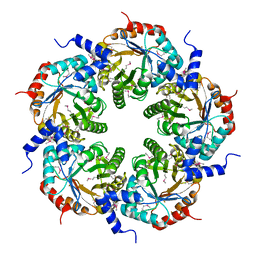 | | Crystal Structure of Cgl0159 From Corynebacterium glutamicum (Brevibacterium flavum). Northeast Structural Genomics Target CgR115 | | Descriptor: | uncharacterized protein Cgl0159 | | Authors: | Seetharaman, J, Neely, H, Wang, H, Janjua, H, Foote, E.L, Xiao, R, Everett, J.K, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-30 | | Release date: | 2009-01-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Cgl0159 From Corynebacterium glutamicum (Brevibacterium flavum). Northeast Structural Genomics Target CgR115
To be Published
|
|
6EM5
 
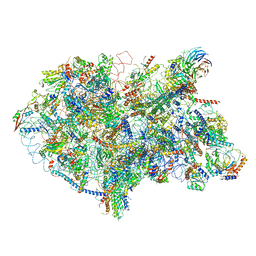 | | State D architectural model (Nsa1-TAP Flag-Ytm1) - Visualizing the assembly pathway of nucleolar pre-60S ribosomes | | Descriptor: | 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 25S ribosomal RNA, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, ... | | Authors: | Kater, L, Cheng, J, Barrio-Garcia, C, Hurt, E, Beckmann, R. | | Deposit date: | 2017-10-01 | | Release date: | 2017-12-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Visualizing the Assembly Pathway of Nucleolar Pre-60S Ribosomes.
Cell, 171, 2017
|
|
3EW7
 
 | | Crystal structure of the Lmo0794 protein from Listeria monocytogenes. Northeast Structural Genomics Consortium target LmR162. | | Descriptor: | Lmo0794 protein | | Authors: | Vorobiev, S.M, Abashidze, M, Seetharaman, J, Wang, D, Foote, E.L, Ciccosanti, C, Janjua, H, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-10-14 | | Release date: | 2008-11-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Crystal structure of the Lmo0794 protein from Listeria monocytogenes.
To be Published
|
|
5NF8
 
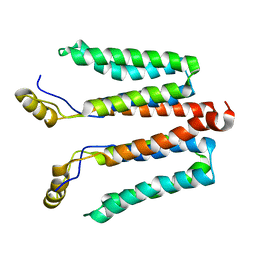 | | Solution structure of detergent-solubilized Rcf1, a yeast mitochondrial inner membrane protein involved in respiratory Complex III/IV supercomplex formation | | Descriptor: | Respiratory supercomplex factor 1, mitochondrial | | Authors: | Zhou, S, Pettersson, P, Sjoholm, J, Sjostrand, D, Hogbom, M, Brzezinski, P, Maler, L, Adelroth, P. | | Deposit date: | 2017-03-13 | | Release date: | 2018-02-28 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of yeast Rcf1, a protein involved in respiratory supercomplex formation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6AKD
 
 | | Crystal structure of IdnL7 | | Descriptor: | '5'-O-(N-(L-ALANYL)-SULFAMOYL)ADENOSINE, AMP-dependent synthetase and ligase, GLYCEROL | | Authors: | Cieslak, J, Miyanaga, A, Kudo, F, Eguchi, T. | | Deposit date: | 2018-08-31 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional and structural characterization of IdnL7, an adenylation enzyme involved in incednine biosynthesis.
Acta Crystallogr F Struct Biol Commun, 75, 2019
|
|
1F4O
 
 | | CRYSTAL STRUCTURE OF GRANCALCIN WITH BOUND CALCIUM | | Descriptor: | CALCIUM ION, GRANCALCIN | | Authors: | Jia, J, Han, Q, Borregaard, N, Lollike, K, Cygler, M. | | Deposit date: | 2000-06-08 | | Release date: | 2000-09-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human grancalcin, a member of the penta-EF-hand protein family.
J.Mol.Biol., 300, 2000
|
|
6EZY
 
 | | ARABIDOPSIS THALIANA GSTF9, GSH AND GSOH BOUND | | Descriptor: | BROMIDE ION, GLUTATHIONE, GLYCEROL, ... | | Authors: | Tossounian, M.A, Wahni, K, VanMolle, I, Rosado, L, Vertommen, D, Messens, J. | | Deposit date: | 2017-11-16 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Redox-regulated methionine oxidation of Arabidopsis thaliana glutathione transferase Phi9 induces H-site flexibility.
Protein Sci., 28, 2019
|
|
6EV1
 
 | | Crystal structure of antibody against schizophyllan | | Descriptor: | Heavy chain, Light chain | | Authors: | Sung, K.H, Josewski, J, Dubel, S, Blankenfeldt, W, Rau, U. | | Deposit date: | 2017-11-01 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.043 Å) | | Cite: | Structural insights into antigen recognition of an anti-beta-(1,6)-beta-(1,3)-D-glucan antibody.
Sci Rep, 8, 2018
|
|
6AME
 
 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 M21A | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-24 | | Release date: | 1999-04-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
6F0G
 
 | | Crystal structure ASF1-ip3 | | Descriptor: | Histone chaperone ASF1A, SULFATE ION, ip3 | | Authors: | Gaubert, A, Guichard, B, Richet, N, Le Du, M.H, Andreani, J, Guerois, R, Ochsenbein, F. | | Deposit date: | 2017-11-20 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design on a Rational Basis of High-Affinity Peptides Inhibiting the Histone Chaperone ASF1.
Cell Chem Biol, 26, 2019
|
|
3EZI
 
 | |
3F05
 
 | |
3FGB
 
 | | Crystal structure of the Q89ZH8_BACTN protein from Bacteroides thetaiotaomicron. Northeast Structural Genomics Consortium target BtR289b. | | Descriptor: | uncharacterized protein Q89ZH8_BACTN | | Authors: | Vorobiev, S.M, Lew, S, Seetharaman, J, Lee, D, Foote, L.E, Ciccosanti, C, Janjua, H, Xiao, R, Acton, T, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-05 | | Release date: | 2008-12-23 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the Q89ZH8_BACTN protein from Bacteroides thetaiotaomicron.
To be Published
|
|
6ADY
 
 | | Crystal structure of the Zika virus NS3 helicase (ADP-Mn2+ complex, form 2) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MANGANESE (II) ION, Serine protease NS3 | | Authors: | Fang, J, Lu, G, Gong, P. | | Deposit date: | 2018-08-02 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic Snapshots of the Zika Virus NS3 Helicase Help Visualize the Reactant Water Replenishment.
ACS Infect Dis, 5, 2019
|
|
6F0F
 
 | | Crystal structure ASF1-ip2_s | | Descriptor: | Histone chaperone ASF1A, ip2_s | | Authors: | Gaubert, A, Guichard, B, Murciano, B, Le Du, M.H, Ochsenbein, F, Guerois, R, Andreani, J. | | Deposit date: | 2017-11-20 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design on a Rational Basis of High-Affinity Peptides Inhibiting the Histone Chaperone ASF1.
Cell Chem Biol, 26, 2019
|
|
6F0K
 
 | | Alternative complex III | | Descriptor: | ActD, ActF, ActH, ... | | Authors: | Sousa, J.S, Calisto, F, Mills, D.J, Pereira, M.M, Vonck, J, Kuehlbrandt, W. | | Deposit date: | 2017-11-20 | | Release date: | 2018-05-09 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Structural basis for energy transduction by respiratory alternative complex III.
Nat Commun, 9, 2018
|
|
