3GFX
 
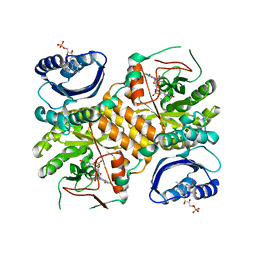 | | Klebsiella pneumoniae BlrP1 pH 4.5 calcium/cy-diGMP complex | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CALCIUM ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Barends, T, Hartmann, E, Griese, J, Beitlich, T, Kirienko, N, Ryjenkov, D, Reinstein, J, Shoeman, R, Gomelsky, M, Schlichting, I. | | Deposit date: | 2009-02-27 | | Release date: | 2009-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and mechanism of a bacterial light-regulated cyclic nucleotide phosphodiesterase.
Nature, 459, 2009
|
|
2HN2
 
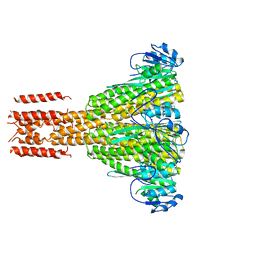 | |
2HNY
 
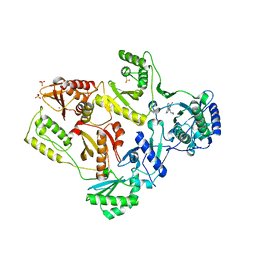 | | Crystal Structure of E138K Mutant HIV-1 Reverse Transcriptase in Complex with Nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Ren, J, Nichols, C.E, Stamp, A, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2006-07-13 | | Release date: | 2006-09-05 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into mechanisms of non-nucleoside drug resistance for HIV-1 reverse transcriptases mutated at codons 101 or 138.
Febs J., 273, 2006
|
|
2VX0
 
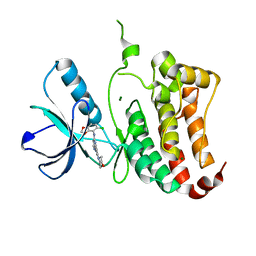 | | ephB4 kinase domain inhibitor complex | | Descriptor: | EPHRIN TYPE-B RECEPTOR 4, MAGNESIUM ION, N'-(5-CHLORO-1,3-BENZODIOXOL-4-YL)-N-(3-MORPHOLIN-4-YLPHENYL)PYRIMIDINE-2,4-DIAMINE | | Authors: | Read, J, Brassington, C.A, Green, I, McCall, E.J, Valentine, A.L, Barratt, D, Leach, A.G, Kettle, J.G. | | Deposit date: | 2008-06-30 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibitors of the Tyrosine Kinase Ephb4. Part 1: Structure-Based Design and Optimization of a Series of 2,4-Bis-Anilinopyrimidines.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3TQT
 
 | | Structure of the D-alanine-D-alanine ligase from Coxiella burnetii | | Descriptor: | D-alanine--D-alanine ligase | | Authors: | Rudolph, M, Cheung, J, Franklin, M.C, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
3TP2
 
 | | Crystal Structure of the Splicing Factor Cwc2 from yeast | | Descriptor: | Pre-mRNA-splicing factor CWC2, SODIUM ION, ZINC ION | | Authors: | Schmitzova, J. | | Deposit date: | 2011-09-07 | | Release date: | 2012-03-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Cwc2 reveals a novel architecture of a multipartite RNA-binding protein.
Embo J., 31, 2012
|
|
3TRB
 
 | | Structure of an addiction module antidote protein of a HigA (higA) family from Coxiella burnetii | | Descriptor: | Virulence-associated protein I | | Authors: | Cheung, J, Franklin, M.C, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
5VRE
 
 | |
3TQN
 
 | | Structure of the transcriptional regulator of the GntR family, from Coxiella burnetii. | | Descriptor: | Transcriptional regulator, GntR family | | Authors: | Rudolph, M, Cheung, J, Franklin, M.C, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-28 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
3TQZ
 
 | | Structure of a deoxyuridine 5'-triphosphate nucleotidohydrolase (dut) from Coxiella burnetii | | Descriptor: | Deoxyuridine 5'-triphosphate nucleotidohydrolase, SULFATE ION | | Authors: | Cheung, J, Franklin, M, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
3TR6
 
 | | Structure of a O-methyltransferase from Coxiella burnetii | | Descriptor: | NICKEL (II) ION, O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Cheung, J, Franklin, M.C, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-28 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
3TR9
 
 | | Structure of a dihydropteroate synthase (folP) in complex with pteroic acid from Coxiella burnetii | | Descriptor: | CHLORIDE ION, Dihydropteroate synthase, PTEROIC ACID, ... | | Authors: | Cheung, J, Franklin, M.C, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
2VWX
 
 | | ephB4 kinase domain inhibitor complex | | Descriptor: | 3-({4-[(5-chloro-1,3-benzodioxol-4-yl)amino]pyrimidin-2-yl}amino)benzenesulfonamide, EPHRIN TYPE-B RECEPTOR 4, MAGNESIUM ION | | Authors: | Read, J, Brassington, C.A, Green, I, McCall, E.J, Valentine, A.L, Barratt, D, Leach, A.G, Kettle, J.G. | | Deposit date: | 2008-06-27 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Inhibitors of the Tyrosine Kinase Ephb4. Part 2: Structure-Based Discovery and Optimisation of 3,5-Bis Substituted Anilinopyrimidines.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2HND
 
 | | Crystal Structure of K101E Mutant HIV-1 Reverse Transcriptase in Complex with Nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Ren, J, Nichols, C.E, Stamp, A, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2006-07-12 | | Release date: | 2006-09-05 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into mechanisms of non-nucleoside drug resistance for HIV-1 reverse transcriptases mutated at codons 101 or 138.
Febs J., 273, 2006
|
|
2HSE
 
 | |
2HGS
 
 | | HUMAN GLUTATHIONE SYNTHETASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLUTATHIONE, MAGNESIUM ION, ... | | Authors: | Polekhina, G, Board, P, Rossjohn, J, Parker, M.W. | | Deposit date: | 1999-01-04 | | Release date: | 1999-06-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular basis of glutathione synthetase deficiency and a rare gene permutation event.
EMBO J., 18, 1999
|
|
5VUD
 
 | | HLA-B*57:01 presenting LSSPVTKSW | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-57 alpha chain, ... | | Authors: | Pymm, P, Rossjohn, J, Vivian, J.P. | | Deposit date: | 2017-05-19 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | HLA-B57 micropolymorphism defines the sequence and conformational breadth of the immunopeptidome.
Nat Commun, 9, 2018
|
|
6ENE
 
 | | OmpF orthologue from Enterobacter cloacae (OmpE35) | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Outer membrane protein (Porin), SULFATE ION, ... | | Authors: | van den Berg, B, Abellon-Ruiz, J, Basle, A. | | Deposit date: | 2017-10-04 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Getting Drugs into Gram-Negative Bacteria: Rational Rules for Permeation through General Porins.
Acs Infect Dis., 4, 2018
|
|
2VWZ
 
 | | ephB4 kinase domain inhibitor complex | | Descriptor: | EPHRIN TYPE-B RECEPTOR 4, MAGNESIUM ION, N-[3-[[4-[(5-CHLORO-1,3-BENZODIOXOL-4-YL)AMINO]PYRIMIDIN-2-YL]AMINO]PHENYL]METHANESULFONAMIDE | | Authors: | Read, J, Brassington, C.A, Green, I, McCall, E.J, Valentine, A.L, Barratt, D, Leach, A.G, Kettle, J.G. | | Deposit date: | 2008-06-30 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Inhibitors of the Tyrosine Kinase Ephb4. Part 2: Structure-Based Discovery and Optimisation of 3,5-Bis Substituted Anilinopyrimidines.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2VX1
 
 | | ephB4 kinase domain inhibitor complex | | Descriptor: | 3-({4-[(5-CHLORO-1,3-BENZODIOXOL-4-YL)AMINO]PYRIMIDIN-2-YL}AMINO)BENZAMIDE, EPHRIN TYPE-B RECEPTOR 4, MAGNESIUM ION | | Authors: | Read, J, Brassington, C.A, Green, I, McCall, E.J, Valentine, A.L, Barratt, D, Leach, A.G, Kettle, J.G. | | Deposit date: | 2008-06-30 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Inhibitors of the Tyrosine Kinase Ephb4. Part 2: Structure-Based Discovery and Optimisation of 3,5-Bis Substituted Anilinopyrimidines.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
1PAL
 
 | | IONIC INTERACTIONS WITH PARVALBUMINS. CRYSTAL STRUCTURE DETERMINATION OF PIKE 4.10 PARVALBUMIN IN FOUR DIFFERENT IONIC ENVIRONMENTS | | Descriptor: | AMMONIUM ION, CALCIUM ION, PARVALBUMIN | | Authors: | Declercq, J.P, Tinant, B, Parello, J, Rambaud, J. | | Deposit date: | 1990-11-08 | | Release date: | 1992-01-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Ionic interactions with parvalbumins. Crystal structure determination of pike 4.10 parvalbumin in four different ionic environments.
J.Mol.Biol., 220, 1991
|
|
5W1O
 
 | | Crystal Structure of HPV16 L1 Pentamer Bound to Heparin Oligosaccharides | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-alpha-D-glucopyranose, 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-alpha-D-glucopyranose, Major capsid protein L1 | | Authors: | Dasgupta, J, Chen, X.S. | | Deposit date: | 2017-06-04 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of oligosaccharide receptor recognition by human papillomavirus.
J. Biol. Chem., 286, 2011
|
|
6GZR
 
 | | Solution NMR structure of the tetramethylrhodamine (TMR) aptamer 3 in complex with 5-TAMRA | | Descriptor: | 5-carboxy methylrhodamine, tetramethylrhodamine aptamer | | Authors: | Duchardt-Ferner, E, Ohlenschlager, O, Kreutz, C.R, Wohnert, J. | | Deposit date: | 2018-07-05 | | Release date: | 2019-07-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of an RNA aptamer in complex with the fluorophore tetramethylrhodamine.
Nucleic Acids Res., 48, 2020
|
|
5W6A
 
 | | HLA-C*06:02 presenting ARTELYRSL | | Descriptor: | ALA-ARG-THR-GLU-LEU-TYR-ARG-SER-LEU, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Mobbs, J.I, Vivian, J.P, Rossjohn, J. | | Deposit date: | 2017-06-16 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The molecular basis for peptide repertoire selection in the human leucocyte antigen (HLA) C*06:02 molecule.
J. Biol. Chem., 292, 2017
|
|
5VJE
 
 | | Class II fructose-1,6-bisphosphate aldolase of Escherichia coli with D-glucitol 1,6-bisphosphate | | Descriptor: | D-Glucitol-1,6-bisphosphate, Fructose-bisphosphate aldolase class 2, SODIUM ION | | Authors: | Coincon, M, Sygusch, J. | | Deposit date: | 2017-04-19 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Active site remodeling during the catalytic cycle in metal-dependent fructose-1,6-bisphosphate aldolases.
J. Biol. Chem., 293, 2018
|
|
