5DQU
 
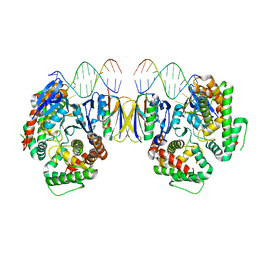 | | Crystal Structure of Cas-DNA-10 complex | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2, DNA (5'-D(*GP*AP*GP*TP*CP*GP*AP*TP*GP*CP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Wang, J, Li, J, Zhao, H, Sheng, G, Wang, M, Yin, M, Wang, Y. | | Deposit date: | 2015-09-15 | | Release date: | 2015-11-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Structural and Mechanistic Basis of PAM-Dependent Spacer Acquisition in CRISPR-Cas Systems.
Cell, 163, 2015
|
|
7RCO
 
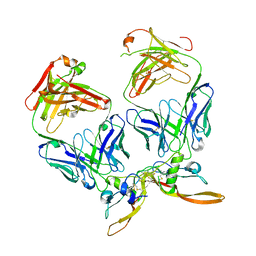 | |
5DUL
 
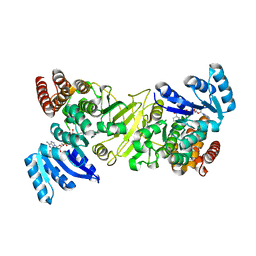 | | 1-deoxy-D-xylulose 5-phosphate reductoisomerase from Yersinia pestis in complex with NADPH | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Osipiuk, J, Mulligan, R, Stam, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-09-18 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase from Yersinia pestis in complex with NADPH .
to be published
|
|
6KOE
 
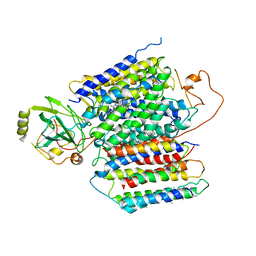 | | X-ray Structure of the proton-pumping cytochrome aa3-600 menaquinol oxidase from Bacillus subtilis | | Descriptor: | 2-HEPTYL-4-HYDROXY QUINOLINE N-OXIDE, AA3-600 quinol oxidase subunit I, AA3-600 quinol oxidase subunit IIII, ... | | Authors: | Xu, J, Ding, Z, Liu, B, Li, J, Gennis, R.B, Zhu, J. | | Deposit date: | 2019-08-09 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | Structure of the cytochromeaa3-600 heme-copper menaquinol oxidase bound to inhibitor HQNO shows TM0 is part of the quinol binding site.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5DKE
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in complex with a 3-naphthyl-substituted, methyl, cis-diaryl-ethylene compound 4,4'-[2-(naphthalen-2-yl)prop-1-ene-1,1-diyl]diphenol | | Descriptor: | 4,4'-[2-(naphthalen-2-yl)prop-1-ene-1,1-diyl]diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-03 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
5DL4
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in complex with a phenylamino-substituted, methyl, triaryl-ethylene derivative 4,4'-{2-[3-(phenylamino)phenyl]prop-1-ene-1,1-diyl}diphenol | | Descriptor: | 4,4'-{2-[3-(phenylamino)phenyl]prop-1-ene-1,1-diyl}diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-04 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
5DYD
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in Complex with the Cyclofenil Derivative 4,4'-{[(3S)-3-(methylsulfanyl)cyclohexylidene]methanediyl}diphenol | | Descriptor: | 4,4'-{[(3S)-3-(methylsulfanyl)cyclohexylidene]methanediyl}diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-24 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.485 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
5DZ3
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in Complex with the Cyclofenil Derivative 4,4'-{[4-(fluoromethyl)cyclohexylidene]methanediyl}diphenol | | Descriptor: | 4,4'-{[4-(fluoromethyl)cyclohexylidene]methanediyl}diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-25 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
6KOC
 
 | | X-ray Structure of the proton-pumping cytochrome aa3-600 menaquinol oxidase from Bacillus subtilis complexed with 3-iodo-N-oxo-2-heptyl-4-hydroxyquinoline | | Descriptor: | 2-heptyl-3-iodanyl-1-oxidanyl-quinolin-4-one, AA3-600 quinol oxidase subunit I, AA3-600 quinol oxidase subunit IIII, ... | | Authors: | Xu, J, Ding, Z, Liu, B, Li, J, Gennis, R.B, Zhu, J. | | Deposit date: | 2019-08-09 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of the cytochromeaa3-600 heme-copper menaquinol oxidase bound to inhibitor HQNO shows TM0 is part of the quinol binding site.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5E1C
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in Complex with the Cyclofenil Derivative dimethyl {(1S)-3-[bis(4-hydroxyphenyl)methylidene]cyclohexyl}propanedioate | | Descriptor: | Estrogen receptor, Nuclear receptor coactivator 2, dimethyl {(1S)-3-[bis(4-hydroxyphenyl)methylidene]cyclohexyl}propanedioate | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-29 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
5DTV
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in complex with a dimethyl-substituted, 3,4-diarylthiophene dioxide core ligand | | Descriptor: | 3,4-bis(4-hydroxy-2-methylphenyl)-1H-1lambda~6~-thiophene-1,1-dione, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-18 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
5E7F
 
 | | Complex between lactococcal phage Tuc2009 RBP head domain and a nanobody (L06) | | Descriptor: | Major structural protein 1, nanobody L06 | | Authors: | Legrand, P, Collins, B, Blangy, S, Murphy, J, Spinelli, S, Gutierrez, C, Richet, N, Kellenberger, C, Desmyter, A, Mahony, J, van Sinderen, D, Cambillau, C. | | Deposit date: | 2015-10-12 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Atomic Structure of the Phage Tuc2009 Baseplate Tripod Suggests that Host Recognition Involves Two Different Carbohydrate Binding Modules.
Mbio, 7, 2016
|
|
5E7T
 
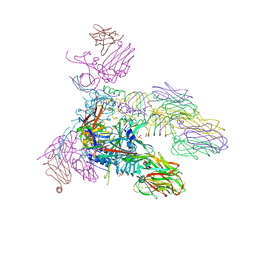 | | Structure of the tripod (BppUct-A-L) from the baseplate of bacteriophage Tuc2009 | | Descriptor: | CALCIUM ION, Major structural protein 1, Minor structural protein 4, ... | | Authors: | Legrand, P, Collins, B, Blangy, S, Murphy, J, Spinelli, S, Gutierrez, C, Richet, N, Kellenberger, C, Desmyter, A, Mahony, J, van Sinderen, D, Cambillau, C. | | Deposit date: | 2015-10-13 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Atomic Structure of the Phage Tuc2009 Baseplate Tripod Suggests that Host Recognition Involves Two Different Carbohydrate Binding Modules.
Mbio, 7, 2016
|
|
5DRM
 
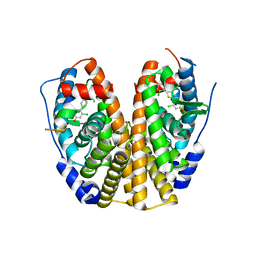 | | Crystal Structure of the ER-alpha Ligand-binding Domain in complex with a dichloro-substituted, 2,5-diarylthiophene-core ligand 4,4'-thiene-2,5-diylbis(3-chlorophenol) | | Descriptor: | 4,4'-thiene-2,5-diylbis(3-chlorophenol), Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-16 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.241 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
5DWE
 
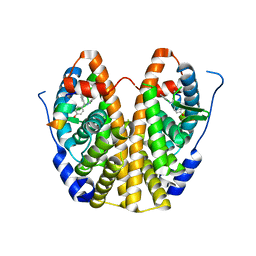 | | Crystal Structure of the ER-alpha Ligand-binding Domain in Complex with a 2-Chloro-substituted Triaryl-imine analog 4,4'-[(2-chlorophenyl)carbonimidoyl]diphenol | | Descriptor: | 4,4'-[(2-chlorophenyl)carbonimidoyl]diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Wright, N.J, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-22 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
6KOB
 
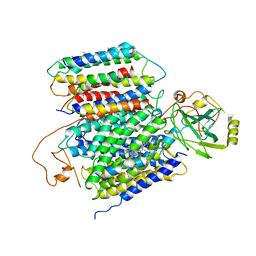 | | X-ray Structure of the proton-pumping cytochrome aa3-600 menaquinol oxidase from Bacillus subtilis | | Descriptor: | AA3-600 quinol oxidase subunit I, AA3-600 quinol oxidase subunit IIII, AA3-600 quinol oxidase subunit IV,Quinol oxidase subunit 4, ... | | Authors: | Xu, J, Ding, Z, Liu, B, Li, J, Gennis, R.B, Zhu, J. | | Deposit date: | 2019-08-09 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of the cytochromeaa3-600 heme-copper menaquinol oxidase bound to inhibitor HQNO shows TM0 is part of the quinol binding site.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3TPI
 
 | | THE GEOMETRY OF THE REACTIVE SITE AND OF THE PEPTIDE GROUPS IN TRYPSIN, TRYPSINOGEN AND ITS COMPLEXES WITH INHIBITORS | | Descriptor: | BOVINE PANCREATIC TRYPSIN INHIBITOR, CALCIUM ION, ISOLEUCINE, ... | | Authors: | Huber, R, Bode, W, Deisenhofer, J, Schwager, P. | | Deposit date: | 1982-09-27 | | Release date: | 1983-01-18 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Geometry of the Reactive Site and of the Peptide Groups in Trypsin, Trypsinogen and its Complexes with Inhibitors
Acta Crystallogr.,Sect.B, 39, 1983
|
|
3UDN
 
 | | Crystal Structure of BACE with Compound 9 | | Descriptor: | 1,2-ETHANEDIOL, 4-cyanobenzyl (3S,5'R)-2-oxo-1,2-dihydrospiro[indole-3,3'-pyrrolidine]-5'-carboxylate, Beta-secretase 1, ... | | Authors: | Efremov, I.V, Vajdos, F.F, Borzilleri, K, Capetta, S, Dorff, P, Dutra, J, Mansour, M, Oborski, C, O'Connell, T, O'Sullivan, T.J, Pandit, J, Wang, H, Withka, J. | | Deposit date: | 2011-10-28 | | Release date: | 2012-04-18 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2.193 Å) | | Cite: | Discovery and optimization of a novel spiropyrrolidine inhibitor of {beta}-secretase (BACE1) through fragment-based drug design.
J.Med.Chem., 55, 2012
|
|
6D55
 
 | | Ras:SOS:Ras in complex with a small molecule activator | | Descriptor: | 6-chloro-2-(2,6-diazaspiro[3.3]heptan-2-yl)-1-[(4-fluoro-3,5-dimethylphenyl)methyl]-4-(4-methylpiperazin-1-yl)-1H-benzimidazole, FORMIC ACID, GLYCEROL, ... | | Authors: | Phan, J, Hodges, T, Fesik, S.W. | | Deposit date: | 2018-04-19 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Discovery and Structure-Based Optimization of Benzimidazole-Derived Activators of SOS1-Mediated Nucleotide Exchange on RAS.
J. Med. Chem., 61, 2018
|
|
3UDK
 
 | | Crystal Structure of BACE with Compound 6 | | Descriptor: | 1,2-ETHANEDIOL, Beta-secretase 1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Efremov, I.V, Vajdos, F.F, Borzilleri, K, Capetta, S, Dorff, P, Dutra, J, Mansour, M, Oborski, C, O'Connell, T, O'Sullivan, T.J, Pandit, J, Wang, H, Withka, J. | | Deposit date: | 2011-10-28 | | Release date: | 2012-04-18 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Discovery and optimization of a novel spiropyrrolidine inhibitor of {beta}-secretase (BACE1) through fragment-based drug design.
J.Med.Chem., 55, 2012
|
|
1MQ4
 
 | | Crystal Structure of Aurora-A Protein Kinase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, AURORA-RELATED KINASE 1, MAGNESIUM ION, ... | | Authors: | Nowakowski, J, Cronin, C.N, McRee, D.E, Knuth, M.W, Nelson, C, Pavletich, N.P, Rodgers, J, Sang, B.-C, Scheibe, D.N, Swanson, R.V, Thompson, D.A. | | Deposit date: | 2002-09-13 | | Release date: | 2003-09-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of the Cancer-Related Aurora-A, FAK and EphA2 Protein Kinases from Nanovolume Crystallography
Structure, 10, 2002
|
|
3UDY
 
 | | Crystal Structure of BACE with Compound 11 | | Descriptor: | 1,2-ETHANEDIOL, 3-bromo-4-cyanobenzyl (3S,5'R)-2-oxo-1,2-dihydrospiro[indole-3,3'-pyrrolidine]-5'-carboxylate, Beta-secretase 1, ... | | Authors: | Efremov, I.V, Vajdos, F.F, Borzilleri, K, Capetta, S, Dorff, P, Dutra, J, Mansour, M, Oborski, C, O'Connell, T, O'Sullivan, T.J, Pandit, J, Wang, H, Withka, J. | | Deposit date: | 2011-10-28 | | Release date: | 2012-04-18 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and optimization of a novel spiropyrrolidine inhibitor of {beta}-secretase (BACE1) through fragment-based drug design.
J.Med.Chem., 55, 2012
|
|
5K54
 
 | | Human muscle fructose-1,6-bisphosphatase E69Q mutant in active R-state | | Descriptor: | Fructose-1,6-bisphosphatase isozyme 2 | | Authors: | Barciszewski, J, Wisniewski, J, Kolodziejczyk, R, Dzugaj, A, Jaskolski, M, Rakus, D. | | Deposit date: | 2016-05-23 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.717 Å) | | Cite: | Structural studies of human muscle FBPase
To Be Published
|
|
6D5G
 
 | | Ras:SOS:Ras in complex with a small molecule activator | | Descriptor: | 6-chloro-1-[(4-fluoro-3,5-dimethylphenyl)methyl]-2-(piperazin-1-yl)-4-(1,2,3,6-tetrahydropyridin-4-yl)-1H-benzimidazole, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Phan, J, Hodges, T, Fesik, S.W. | | Deposit date: | 2018-04-19 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Discovery and Structure-Based Optimization of Benzimidazole-Derived Activators of SOS1-Mediated Nucleotide Exchange on RAS.
J. Med. Chem., 61, 2018
|
|
6D5V
 
 | | Ras:SOS:Ras in complex with a small molecule activator | | Descriptor: | 1-[(3-chloro-4-fluorophenyl)methyl]-5,6-dimethyl-1H-benzimidazol-2-amine, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Phan, J, Hodges, T, Fesik, S.W. | | Deposit date: | 2018-04-19 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Discovery and Structure-Based Optimization of Benzimidazole-Derived Activators of SOS1-Mediated Nucleotide Exchange on RAS.
J. Med. Chem., 61, 2018
|
|
