4QOE
 
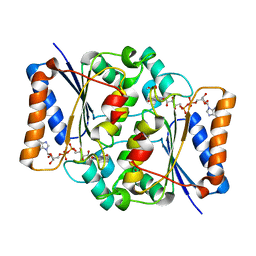 | | The value 'crystal structure of fad quinone reductase 2 at 1.45A | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | 著者 | Serriere, J, Boutin, J.A, Isabet, T, Antoine, M, Ferry, G. | | 登録日 | 2014-06-20 | | 公開日 | 2015-07-01 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | The value 'crystal structure of fad quinone reductase 2 at 1.45A
To be Published
|
|
3P9M
 
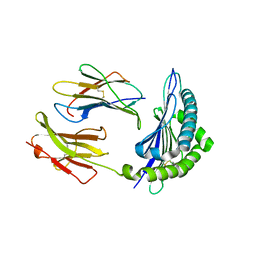 | | Crystal Structure of H2-Kb in complex with a mutant of the chicken ovalbumin epitope OVA-G4 | | 分子名称: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, K-B alpha chain, ... | | 著者 | Wesselingh, R, Gras, S, Guillonneau, C, Turner, S.J, Rossjohn, J. | | 登録日 | 2010-10-18 | | 公開日 | 2011-10-19 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Affinity thresholds for naive CD8+ CTL activation by peptides and engineered influenza A viruses
J.Immunol., 187, 2011
|
|
8HDA
 
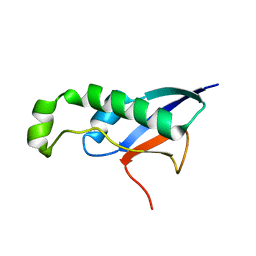 | |
5G5L
 
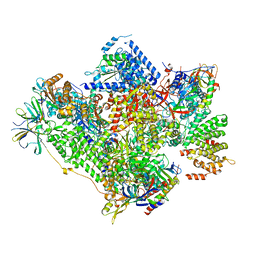 | | RNA polymerase I-Rrn3 complex at 4.8 A resolution | | 分子名称: | DNA-DIRECTED RNA POLYMERASE I SUBUNIT RPA12, DNA-DIRECTED RNA POLYMERASE I SUBUNIT RPA135, DNA-DIRECTED RNA POLYMERASE I SUBUNIT RPA14, ... | | 著者 | Engel, C, Plitzko, J, Cramer, P. | | 登録日 | 2016-05-26 | | 公開日 | 2016-07-27 | | 最終更新日 | 2018-10-24 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | RNA Polymerase I-Rrn3 Complex at 4.8 A Resolution
Nat.Commun., 7, 2016
|
|
1Y6V
 
 | | Structure of E. coli Alkaline Phosphatase in presence of cobalt at 1.60 A resolution | | 分子名称: | Alkaline phosphatase, COBALT (II) ION, PHOSPHATE ION, ... | | 著者 | Wang, J, Stieglitz, K, Kantrowitz, E.R. | | 登録日 | 2004-12-07 | | 公開日 | 2005-06-21 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Metal Specificity Is Correlated with Two Crucial Active Site Residues in Escherichia coli Alkaline Phosphatase(,).
Biochemistry, 44, 2005
|
|
4QOF
 
 | | Crystal structure of fmn quinone reductase 2 AT 1.55A | | 分子名称: | FLAVIN MONONUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ZINC ION | | 著者 | Serriere, J, Boutin, J.A, Isabet, T, Antoine, M, Ferry, G. | | 登録日 | 2014-06-20 | | 公開日 | 2015-07-01 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Crystal structure of FMN quinone reductase 2 at 1.55A
To be Published
|
|
5G3Y
 
 | | Crystal structure of adenylate kinase ancestor 1 with Zn and ADP bound | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENYLATE KINSE, ZINC ION | | 著者 | Nguyen, V, Kutter, S, English, J, Kern, D. | | 登録日 | 2016-05-03 | | 公開日 | 2016-12-28 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.18 Å) | | 主引用文献 | Evolutionary drivers of thermoadaptation in enzyme catalysis.
Science, 355, 2017
|
|
5G4N
 
 | | Crystal structure of the p53 cancer mutant Y220C in complex with a difluorinated derivative of the small molecule stabilizer Phikan083 | | 分子名称: | 1-[9-(2,2-difluoroethyl)-9H-carbazol-3-yl]-N-methylmethanamine, CELLULAR TUMOR ANTIGEN P53, GLYCEROL, ... | | 著者 | Joerger, A.C, Bauer, M, Jones, R.N, Spencer, J. | | 登録日 | 2016-05-13 | | 公開日 | 2016-06-22 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Harnessing Fluorine-Sulfur Contacts and Multipolar Interactions for the Design of P53 Mutant Y220C Rescue Drugs.
Acs Chem.Biol., 11, 2016
|
|
5EPJ
 
 | | Crystal Structure of chromodomain of CBX7 in complex with inhibitor UNC3866 | | 分子名称: | Chromobox protein homolog 7, UNKNOWN ATOM OR ION, peptide-like inhibitor UNC3866 | | 著者 | Liu, Y, Tempel, W, Walker, J.R, Stuckey, J.I, Dickson, B.M, James, L.I, Frye, S.V, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2015-11-11 | | 公開日 | 2015-12-16 | | 最終更新日 | 2019-11-27 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal Structure of chromodomain of CBX7 in complex with inhibitor UNC3866
to be published
|
|
4QPJ
 
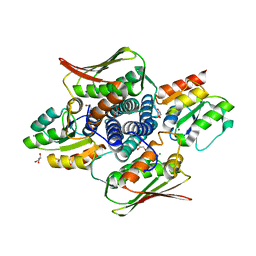 | | 2.7 Angstrom Structure of a Phosphotransferase in Complex with a Receiver Domain | | 分子名称: | CALCIUM ION, CHLORIDE ION, Cell cycle response regulator CtrA, ... | | 著者 | Willett, J.W, Herrou, J, Crosson, S. | | 登録日 | 2014-06-23 | | 公開日 | 2015-07-15 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.742 Å) | | 主引用文献 | Structural asymmetry in a conserved signaling system that regulates division, replication, and virulence of an intracellular pathogen.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5EKY
 
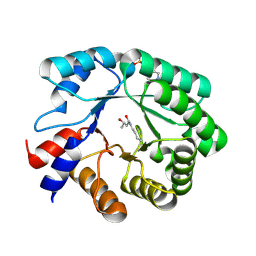 | | Crystal structure of deoxyribose-phosphate aldolase from Escherichia coli (K58E-Y96W mutant) | | 分子名称: | 1,3-BUTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Deoxyribose-phosphate aldolase | | 著者 | Classen, T, Dick, M, Pietruszka, J, Weiergraeber, O.H. | | 登録日 | 2015-11-04 | | 公開日 | 2016-05-04 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Mechanism-based inhibition of an aldolase at high concentrations of its natural substrate acetaldehyde: structural insights and protective strategies.
Chem Sci, 7, 2016
|
|
5ERP
 
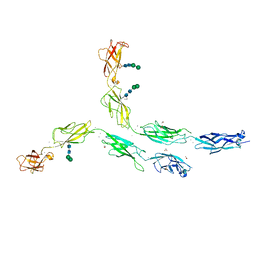 | | Crystal structure of human Desmocollin-2 ectodomain fragment EC2-5 | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | 著者 | Harrison, O.J, Brasch, J, Shapiro, L. | | 登録日 | 2015-11-14 | | 公開日 | 2016-06-15 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural basis of adhesive binding by desmocollins and desmogleins.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1YGS
 
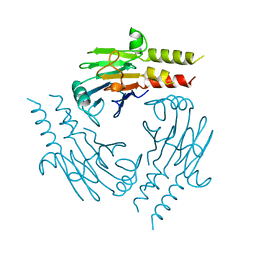 | | CRYSTAL STRUCTURE OF THE SMAD4 TUMOR SUPPRESSOR C-TERMINAL DOMAIN | | 分子名称: | SMAD4 | | 著者 | Shi, Y, Hata, A, Lo, R.S, Massague, J, Pavletich, N.P. | | 登録日 | 1997-10-03 | | 公開日 | 1998-07-08 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | A structural basis for mutational inactivation of the tumour suppressor Smad4.
Nature, 388, 1997
|
|
1YHF
 
 | | Crystal Structure of Conserved SPY1581 Protein of Unknown Function from Streptococcus pyogenes | | 分子名称: | hypothetical protein SPy1581 | | 著者 | Osipiuk, J, Lezondra, L, Moy, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2005-01-07 | | 公開日 | 2005-02-22 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | X-ray crystal structure of conserved hypothetical SPy1581 protein from Streptococcus pyogenes.
To be Published
|
|
4QPF
 
 | | New lower bone affinity bisphosphonate drug design for effective use in diseases characterized by abnormal bone resorption | | 分子名称: | Farnesyl pyrophosphate synthase, MAGNESIUM ION, [1-fluoro-2-(imidazo[1,2-a]pyridin-3-yl)ethane-1,1-diyl]bis(phosphonic acid) | | 著者 | Ebetino, F.H, Lundy, M, Kwaasi, A.A, Dunford, J.E, Duan, Z, Triffitt, J, Mazur, A, Jeans, G, Barnett, B.L, Russell, R.G.G. | | 登録日 | 2014-06-23 | | 公開日 | 2015-06-24 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | New lower bone affinity bisphosphonate drug design for effective use in diseases characterized by abnormal bone resorption
To be Published
|
|
1YHZ
 
 | | Crystal structure of Arabidopsis thaliana Acetohydroxyacid synthase In Complex With A Sulfonylurea Herbicide, Chlorsulfuron | | 分子名称: | 1-(2-CHLOROPHENYLSULFONYL)-3-(4-METHOXY-6-METHYL-L,3,5-TRIAZIN-2-YL)UREA, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Acetolactate synthase, ... | | 著者 | McCourt, J.A, Pang, S.S, King-Scott, J, Guddat, L.W, Duggleby, R.G. | | 登録日 | 2005-01-10 | | 公開日 | 2006-01-17 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Herbicide-binding sites revealed in the structure of plant acetohydroxyacid synthase
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1Y8F
 
 | |
1YA0
 
 | | Crystal structure of the N-terminal domain of human SMG7 | | 分子名称: | SMG-7 transcript variant 2, SULFATE ION | | 著者 | Fukuhara, N, Ebert, J, Unterholzner, L, Lindner, D, Izaurralde, E, Conti, E. | | 登録日 | 2004-12-17 | | 公開日 | 2005-03-15 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | SMG7 Is a 14-3-3-like Adaptor in the Nonsense-Mediated mRNA Decay Pathway.
Mol.Cell, 17, 2005
|
|
3PAB
 
 | | Crystal Structure of H2-Kb in complex with a mutant of the chicken ovalbumin epitope OVA-E1 | | 分子名称: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, K-B alpha chain, ... | | 著者 | Wesselingh, R, Gras, S, Guillonneau, C, Turner, S.J, Rossjohn, J. | | 登録日 | 2010-10-19 | | 公開日 | 2011-10-19 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Affinity thresholds for naive CD8+ CTL activation by peptides and engineered influenza A viruses
J.Immunol., 187, 2011
|
|
5EDN
 
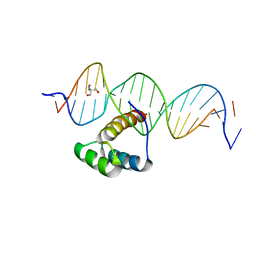 | | Structure of HOXB13-DNA(TCG) complex | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA (5'-D(P*GP*GP*AP*CP*CP*TP*CP*GP*TP*AP*AP*AP*AP*CP*AP*CP*AP*AP*C)-3'), DNA (5'-D(P*GP*TP*TP*GP*TP*GP*TP*TP*TP*TP*AP*CP*GP*AP*GP*GP*TP*CP*C)-3'), ... | | 著者 | Morgunova, E, Yin, Y, Jolma, A, Popov, A, Taipale, J. | | 登録日 | 2015-10-21 | | 公開日 | 2016-11-09 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Two distinct DNA sequences recognized by transcription factors represent enthalpy and entropy optima.
Elife, 7, 2018
|
|
1YCD
 
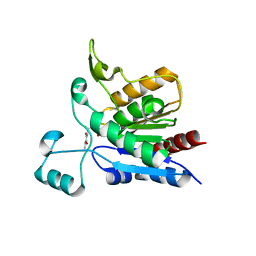 | | Crystal structure of yeast FSH1/YHR049W, a member of the serine hydrolase family | | 分子名称: | 2-HYDROXY-4,5-DIOXOHEPTYL HYDROGEN PHOSPHONATE, Hypothetical 27.3 kDa protein in AAP1-SMF2 intergenic region | | 著者 | Leulliot, N, Graille, M, Coste, F, Quevillon-Cheruel, S, Janin, J, van Tilbeurgh, H, Paris-Sud Yeast Structural Genomics (YSG) | | 登録日 | 2004-12-22 | | 公開日 | 2005-05-10 | | 最終更新日 | 2021-10-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of yeast YHR049W/FSH1, a member of the serine hydrolase family.
Protein Sci., 14, 2005
|
|
153D
 
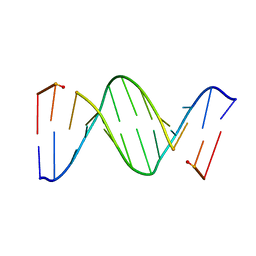 | | CRYSTAL STRUCTURE OF A MISPAIRED DODECAMER, D(CGAGAATTC(O6ME)GCG)2, CONTAINING A CARCINOGENIC O6-METHYLGUANINE | | 分子名称: | DNA (5'-D(*CP*GP*AP*GP*AP*AP*TP*TP*CP*(6OG)P*CP*G)-3') | | 著者 | Ginell, S.L, Vojtechovsky, J, Gaffney, B, Jones, R, Berman, H.M. | | 登録日 | 1993-12-16 | | 公開日 | 1994-01-15 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal structure of a mispaired dodecamer, d(CGAGAATTC(O6Me)GCG)2, containing a carcinogenic O6-methylguanine
Biochemistry, 33, 1994
|
|
1Y2B
 
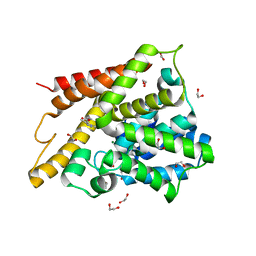 | | Catalytic Domain Of Human Phosphodiesterase 4D In Complex With 3,5-dimethyl-1H-pyrazole-4-carboxylic acid ethyl ester | | 分子名称: | 1,2-ETHANEDIOL, 3,5-DIMETHYL-1H-PYRAZOLE-4-CARBOXYLIC ACID ETHYL ESTER, MAGNESIUM ION, ... | | 著者 | Card, G.L, Blasdel, L, England, B.P, Zhang, C, Suzuki, Y, Gillette, S, Fong, D, Ibrahim, P.N, Artis, D.R, Bollag, G, Milburn, M.V, Kim, S.-H, Schlessinger, J, Zhang, K.Y.J. | | 登録日 | 2004-11-22 | | 公開日 | 2005-03-01 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | A family of phosphodiesterase inhibitors discovered by cocrystallography and scaffold-based drug design
Nat.Biotechnol., 23, 2005
|
|
4QQV
 
 | | Extracellular domains of mouse IL-3 beta receptor | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-3 receptor class 2 subunit beta | | 著者 | Jackson, C.J, Young, I.G, Murphy, J.M, Carr, P.D, Ewens, C.L, Dai, J, Ollis, D.L. | | 登録日 | 2014-06-30 | | 公開日 | 2014-09-03 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.45 Å) | | 主引用文献 | Crystal structure of the mouse interleukin-3 beta-receptor: insights into interleukin-3 binding and receptor activation.
Biochem.J., 463, 2014
|
|
5EM3
 
 | |
