7WMQ
 
 | | Crystal Structure of the second bromodomain of human BRD2 in complex with the inhibitor Y13157 | | Descriptor: | 2-(2-cyclobutyl-1~{H}-imidazol-5-yl)-7-[2-(4-fluoranyl-2,6-dimethyl-phenoxy)-5-(2-oxidanylpropan-2-yl)phenyl]-5-methyl-furo[3,2-c]pyridin-4-one, FORMIC ACID, GLYCEROL, ... | | Authors: | Li, J, Zhang, C, Xu, H, Zhuang, X, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2022-01-16 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure-Based Discovery and Optimization of Furo[3,2- c ]pyridin-4(5 H )-one Derivatives as Potent and Second Bromodomain (BD2)-Selective Bromo and Extra Terminal Domain (BET) Inhibitors.
J.Med.Chem., 65, 2022
|
|
7VZU
 
 | | The structure of GdmN Y82F mutant | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, GdmN, ... | | Authors: | Wei, J, Zheng, J, Zhou, J, Kang, Q, Bai, L. | | Deposit date: | 2021-11-16 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Endowing homodimeric carbamoyltransferase GdmN with iterative functions through structural characterization and mechanistic studies.
Nat Commun, 13, 2022
|
|
1BS2
 
 | | YEAST ARGINYL-TRNA SYNTHETASE | | Descriptor: | ARGININE, PROTEIN (ARGINYL-TRNA SYNTHETASE) | | Authors: | Cavarelli, J, Delagouute, B, Eriani, G, Gangloff, J, Moras, D. | | Deposit date: | 1998-08-31 | | Release date: | 1999-08-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | L-arginine recognition by yeast arginyl-tRNA synthetase.
EMBO J., 17, 1998
|
|
7VZY
 
 | | The structure of GdmN complex with AMP and 20-O-methyl-19-chloroproansamitocin | | Descriptor: | (5~{S},6~{E},8~{S},9~{S},12~{R},15~{E})-21-chloranyl-12,20-dimethoxy-6,8,16-trimethyl-5,9-bis(oxidanyl)-2-azabicyclo[16.3.1]docosa-1(21),6,15,18(22),19-pentaene-3,11-dione, 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Wei, J, Zheng, J, Zhou, J, Kang, Q, Bai, L. | | Deposit date: | 2021-11-17 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Endowing homodimeric carbamoyltransferase GdmN with iterative functions through structural characterization and mechanistic studies.
Nat Commun, 13, 2022
|
|
7VZZ
 
 | | The structure of GdmN in complex with the natural tetrahedral intermediate, carbamoyl adenylate, and 20-O-methyl-19-chloroproansamitocin | | Descriptor: | (5~{S},6~{E},8~{S},9~{S},12~{R},15~{E})-21-chloranyl-12,20-dimethoxy-6,8,16-trimethyl-5,9-bis(oxidanyl)-2-azabicyclo[16.3.1]docosa-1(21),6,15,18(22),19-pentaene-3,11-dione, 1,2-ETHANEDIOL, 5'-O-[(S)-(carbamoyloxy)(hydroxy)phosphoryl]adenosine, ... | | Authors: | Wei, J, Zheng, J, Zhou, J, Kang, Q, Bai, L. | | Deposit date: | 2021-11-17 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Endowing homodimeric carbamoyltransferase GdmN with iterative functions through structural characterization and mechanistic studies.
Nat Commun, 13, 2022
|
|
7W1I
 
 | | Crystal structure of carboxylesterase mutant from Thermobifida fusca with C8X and C9C | | Descriptor: | 4-(2-hydroxyethyloxycarbonyl)benzoic acid, Carboxylesterase, bis(2-hydroxyethyl) benzene-1,4-dicarboxylate | | Authors: | Han, X, Gerlis, H, Li, Z, Gao, J, Wei, R, Liu, W. | | Deposit date: | 2021-11-19 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural Insights into (Tere)phthalate-Ester Hydrolysis by a Carboxylesterase and Its Role in Promoting PET Depolymerization
Acs Catalysis, 12, 2022
|
|
7W1K
 
 | | Crystal structure of carboxylesterase from Thermobifida fusca | | Descriptor: | Carboxylesterase | | Authors: | Han, X, Gerlis, H, Li, Z, Gao, J, Wei, R, Liu, W. | | Deposit date: | 2021-11-19 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural Insights into (Tere)phthalate-Ester Hydrolysis by a Carboxylesterase and Its Role in Promoting PET Depolymerization
Acs Catalysis, 12, 2022
|
|
4IA0
 
 | | Crystal structure of the PDE5A1 catalytic domain in complex with novel inhibitors | | Descriptor: | 5-bromo-2-{2-ethoxy-5-[(4-methylpiperazin-1-yl)sulfonyl]phenyl}-6-octylpyrimidin-4(3H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Ren, J, Chen, T, Xu, Y. | | Deposit date: | 2012-12-05 | | Release date: | 2014-01-01 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Exploration of the 5-bromopyrimidin-4(3H)-ones as potent inhibitors of PDE5.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
8I4Z
 
 | | CalA3 with hydrolysis product | | Descriptor: | 11-oxidanylidene-11-(1~{H}-pyrrol-2-yl)undecanoic acid, Beta-ketoacyl-acyl-carrier-protein synthase I | | Authors: | Wang, J, Wang, Z. | | Deposit date: | 2023-01-21 | | Release date: | 2023-02-22 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | C-N bond formation by a polyketide synthase.
Nat Commun, 14, 2023
|
|
8I4Y
 
 | | CalA3 complex structure with amidation product | | Descriptor: | 11-oxidanylidene-11-(1~{H}-pyrrol-2-yl)undecanoic acid, 3-HYDROXYANTHRANILIC ACID, Beta-ketoacyl-acyl-carrier-protein synthase I | | Authors: | Wang, J, Wang, Z. | | Deposit date: | 2023-01-21 | | Release date: | 2023-02-22 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | C-N bond formation by a polyketide synthase.
Nat Commun, 14, 2023
|
|
6SK6
 
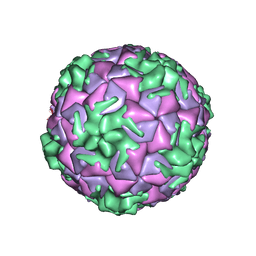 | | Cryo-EM structure of rhinovirus-B5 | | Descriptor: | Rhinovirus B5 VP1, Rhinovirus B5 VP2, Rhinovirus B5 VP3, ... | | Authors: | Wald, J, Goessweiner-Mohr, N, Blaas, D, Pasin, M. | | Deposit date: | 2019-08-14 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of pleconaril-resistant rhinovirus-B5 complexed to the antiviral OBR-5-340 reveals unexpected binding site.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7W1J
 
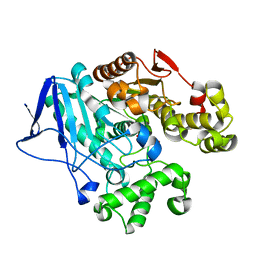 | | Crystal structure of carboxylesterase from Thermobifida fusca with J1K | | Descriptor: | 4-(2-hydroxyethylcarbamoyl)benzoic acid, Carboxylesterase | | Authors: | Han, X, Gerlis, H, Li, Z, Gao, J, Wei, R, Liu, W. | | Deposit date: | 2021-11-19 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural Insights into (Tere)phthalate-Ester Hydrolysis by a Carboxylesterase and Its Role in Promoting PET Depolymerization
Acs Catalysis, 12, 2022
|
|
7W1L
 
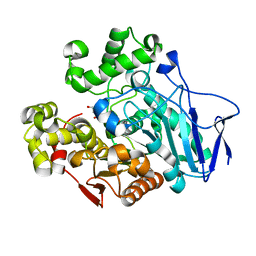 | | Crystal structure of carboxylesterase mutant from Thermobifida fusca with C8X | | Descriptor: | Carboxylesterase, bis(2-hydroxyethyl) benzene-1,4-dicarboxylate | | Authors: | Han, X, Gerlis, H, Li, Z, Gao, J, Wei, R, Liu, W. | | Deposit date: | 2021-11-19 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structural Insights into (Tere)phthalate-Ester Hydrolysis by a Carboxylesterase and Its Role in Promoting PET Depolymerization
Acs Catalysis, 12, 2022
|
|
2C30
 
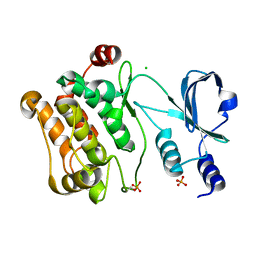 | | Crystal Structure Of The Human P21-Activated Kinase 6 | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, SERINE/THREONINE-PROTEIN KINASE PAK 6 | | Authors: | Filippakopoulos, P, Berridge, G, Bray, J, Burgess, N, Colebrook, S, Das, S, Eswaran, J, Gileadi, O, Papagrigoriou, E, Savitsky, P, Smee, C, Turnbull, A, Sundstrom, M, Arrowsmith, C, Weigelt, J, Edwards, A, von Delft, F, Knapp, S. | | Deposit date: | 2005-10-02 | | Release date: | 2006-02-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of the P21-Activated Kinases Pak4, Pak5, and Pak6 Reveal Catalytic Domain Plasticity of Active Group II Paks.
Structure, 15, 2007
|
|
7NOZ
 
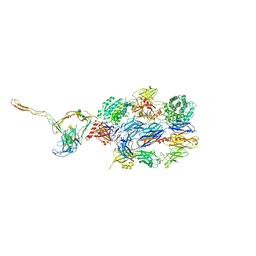 | | Structure of the nanobody stablized properdin bound alternative pathway proconvertase C3b:FB:FP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C3 alpha chain, Complement C3 beta chain, ... | | Authors: | Lorenzen, J, Pedersen, D.V, Andersen, G.R. | | Deposit date: | 2021-02-26 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structure determination of an unstable macromolecular complex enabled by nanobody-peptide bridging.
Protein Sci., 31, 2022
|
|
6PAB
 
 | |
2BI4
 
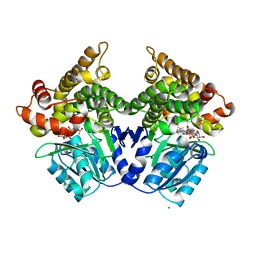 | | Lactaldehyde:1,2-propanediol oxidoreductase of Escherichia coli | | Descriptor: | CHLORIDE ION, FE (III) ION, LACTALDEHYDE REDUCTASE, ... | | Authors: | Montella, C, Bellsolell, L, Badia, J, Baldoma, L, Perez, R, Coll, M, Aguilar, J. | | Deposit date: | 2005-01-20 | | Release date: | 2005-07-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal Structure of an Iron-Dependent Group III Dehydrogenase that Interconverts L-Lactaldehyde and L-1,2-Propanediol in Escherichia Coli
J.Bacteriol., 187, 2005
|
|
7UDH
 
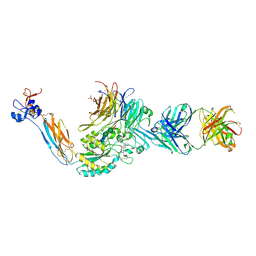 | | Integrin alaphIIBbeta3 complex with BMS4-3 | | Descriptor: | (4-{[(5S)-3-(piperidin-4-yl)-4,5-dihydro-1,2-oxazol-5-yl]methyl}piperazin-1-yl)acetic acid, 10E5 Fab heavy chain, 10E5 Fab light chain, ... | | Authors: | Lin, F.-Y, Zhu, J, Zhu, J, Springer, T.A. | | Deposit date: | 2022-03-19 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.99998844 Å) | | Cite: | A general chemical principle for creating closure-stabilizing integrin inhibitors.
Cell, 185, 2022
|
|
7UK9
 
 | | Integrin alaphIIBbeta3 complex with lamifiban (Mn) | | Descriptor: | 10E5 Fab heavy chain, 10E5 Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lin, F.-Y, Zhu, J, Zhu, J, Springer, T.A. | | Deposit date: | 2022-03-31 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.60001969 Å) | | Cite: | A general chemical principle for creating closure-stabilizing integrin inhibitors.
Cell, 185, 2022
|
|
7UFH
 
 | | Integrin alaphIIBbeta3 complex with fradafiban (Mn/Ca) | | Descriptor: | 10E5 Fab heavy chain, 10E5 Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lin, F.-Y, Zhu, J, Zhu, J, Springer, T.A. | | Deposit date: | 2022-03-22 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.99768162 Å) | | Cite: | A general chemical principle for creating closure-stabilizing integrin inhibitors.
Cell, 185, 2022
|
|
6MIB
 
 | |
7UKT
 
 | | Integrin alaphIIBbeta3 complex with BMS4.2 | | Descriptor: | (1-{[(5S)-3-(4-carbamimidoylphenyl)-4,5-dihydro-1,2-oxazol-5-yl]methyl}piperidin-4-yl)acetic acid, 10E5 Fab heavy chain, 10E5 Fab light chain, ... | | Authors: | Lin, F.-Y, Zhu, J, Zhu, J, Springer, T.A. | | Deposit date: | 2022-04-01 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.36994433 Å) | | Cite: | A general chemical principle for creating closure-stabilizing integrin inhibitors.
Cell, 185, 2022
|
|
7UCY
 
 | | Integrin alaphIIBbeta3 complex with gantofiban | | Descriptor: | (4-{[(5R)-3-(4-carbamimidoylphenyl)-2-oxo-1,3-oxazolidin-5-yl]methyl}piperazin-1-yl)acetic acid, 10E5 Fab heavy chain, 10E5 Fab light chain, ... | | Authors: | Lin, F.-Y, Zhu, J, Zhu, J, Springer, T.A. | | Deposit date: | 2022-03-17 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.34996319 Å) | | Cite: | A general chemical principle for creating closure-stabilizing integrin inhibitors.
Cell, 185, 2022
|
|
7UKO
 
 | | Integrin alaphIIBbeta3 complex with sibrafiban (Mn) | | Descriptor: | 10E5 Fab heavy chain, 10E5 Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lin, F.-Y, Zhu, J, Zhu, J, Springer, T.A. | | Deposit date: | 2022-04-01 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | A general chemical principle for creating closure-stabilizing integrin inhibitors.
Cell, 185, 2022
|
|
6PA6
 
 | |
