3HUO
 
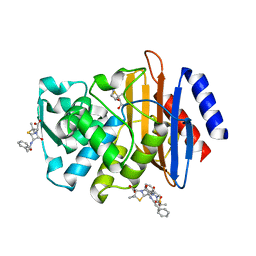 | | X-ray crystallographic structure of CTX-M-9 S70G in complex with benzylpenicillin | | Descriptor: | (2R,4S)-2-{(R)-carboxy[(phenylacetyl)amino]methyl}-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, CTX-M-9 extended-spectrum beta-lactamase, PENICILLIN G | | Authors: | Delmas, J, Leyssene, D, Dubois, D, Robin, F, Bonnet, R. | | Deposit date: | 2009-06-15 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Dynamic view of the early and late steps of the catalytic mechanism mediated by the emerging enzymes CTX-M.
To be Published
|
|
1UHY
 
 | | Crystal structure of d(GCGATAGC): the base-intercalated duplex | | Descriptor: | 5'-D(*GP*(CBR)P*GP*AP*TP*AP*GP*C)-3', CHLORIDE ION, COBALT HEXAMMINE(III), ... | | Authors: | Kondo, J, Umeda, S.I, Fujita, K, Sunami, T, Takenaka, A. | | Deposit date: | 2003-07-13 | | Release date: | 2004-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray analyses of d(GCGAXAGC) containing G and T at X: the base-intercalated duplex is still stable even in point mutants at the fifth residue.
J.Synchrotron Radiat., 11, 2004
|
|
1UI1
 
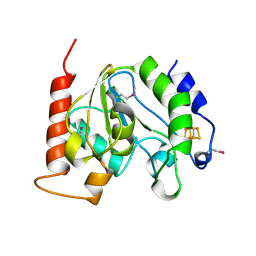 | | Crystal Structure Of Uracil-DNA Glycosylase From Thermus Thermophilus HB8 | | Descriptor: | IRON/SULFUR CLUSTER, Uracil-DNA Glycosylase | | Authors: | Hoseki, J, Okamoto, A, Masui, R, Shibata, T, Inoue, Y, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-14 | | Release date: | 2003-10-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of a Family 4 Uracil-DNA Glycosylase from Thermus thermophilus HB8
J.Mol.Biol., 333, 2003
|
|
4A5X
 
 | | Structures of MITD1 | | Descriptor: | 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, CHARGED MULTIVESICULAR BODY PROTEIN 1A, GLYCEROL, ... | | Authors: | Hadders, M.A, Agromayor, M, Caballe, A, Obita, T, Perisic, O, Williams, R.L, Martin-Serrano, J. | | Deposit date: | 2011-10-28 | | Release date: | 2012-11-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Escrt-III Binding Protein Mitd1 is Involved in Cytokinesis and Has an Unanticipated Pld Fold that Binds Membranes.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4AA9
 
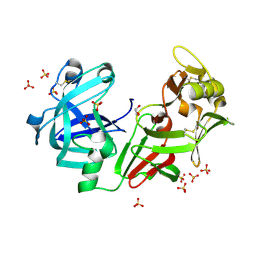 | | Camel chymosin at 1.6A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHYMOSIN, GLYCEROL, ... | | Authors: | Langholm Jensen, J, Molgaard, A, Navarro Poulsen, J.C, van den Brink, J.M, Harboe, M, Simonsen, J.B, Qvist, K.B, Larsen, S. | | Deposit date: | 2011-11-30 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Camel and Bovine Chymosin: The Relationship between Their Structures and Cheese-Making Properties.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3HVZ
 
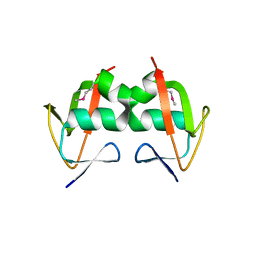 | | Crystal Structure of the TGS domain of the CLOLEP_03100 protein from Clostridium leptum, Northeast Structural Genomics Consortium Target QlR13A | | Descriptor: | Uncharacterized protein | | Authors: | Forouhar, F, Lew, S, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-16 | | Release date: | 2009-06-23 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Northeast Structural Genomics Consortium Target QlR13A
To be Published
|
|
1UHX
 
 | | Crystal structure of d(GCGAGAGC): the base-intercalated duplex | | Descriptor: | 5'-D(*GP*(CBR)P*GP*AP*GP*AP*GP*C)-3', CHLORIDE ION, COBALT HEXAMMINE(III), ... | | Authors: | Kondo, J, Umeda, S.I, Fujita, K, Sunami, T, Takenaka, A. | | Deposit date: | 2003-07-13 | | Release date: | 2004-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray analyses of d(GCGAXAGC) containing G and T at X: the base-intercalated duplex is still stable even in point mutants at the fifth residue.
J.Synchrotron Radiat., 11, 2004
|
|
1UI0
 
 | | Crystal Structure Of Uracil-DNA Glycosylase From Thermus Thermophilus HB8 | | Descriptor: | IRON/SULFUR CLUSTER, SULFATE ION, URACIL, ... | | Authors: | Hoseki, J, Okamoto, A, Masui, R, Shibata, T, Inoue, Y, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-14 | | Release date: | 2003-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of a Family 4 Uracil-DNA Glycosylase from Thermus thermophilus HB8
J.Mol.Biol., 333, 2003
|
|
4AE9
 
 | | Structure and function of the Human Sperm-Specific Isoform of Protein Kinase A (PKA) Catalytic Subunit C alpha 2 | | Descriptor: | CAMP-DEPENDENT PROTEIN KINASE CATALYTIC SUBUNIT ALPHA | | Authors: | Hereng, T.H, Backe, P.H, Kahmann, J, Scheich, C, Bjoras, M, Skalhegg, B.S, Rosendal, K.R. | | Deposit date: | 2012-01-09 | | Release date: | 2012-04-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Function of the Human Sperm-Specific Isoform of Protein Kinase a (Pka) Catalytic Subunit Calpha2
J.Struct.Biol., 178, 2012
|
|
4AFV
 
 | | THE STRUCTURE OF METACASPASE 2 FROM T. BRUCEI DETERMINED IN THE PRESENCE OF CALCIUM CHLORIDE | | Descriptor: | METACASPASE MCA2 | | Authors: | McLuskey, K, Rudolf, J, Isaacs, N.W, Coombs, G.H, Moss, C.X, Mottram, J.C. | | Deposit date: | 2012-01-23 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of a Trypanosoma Brucei Metacaspase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4AME
 
 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 T18A | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-24 | | Release date: | 1999-04-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
3HZE
 
 | | Crystal Structure of Ycf54 protein from Thermosynechococcus elongatus, Northeast Structural Genomics Consortium Target TeR59 | | Descriptor: | PHOSPHATE ION, Ycf54 protein | | Authors: | Kuzin, A, Abashidze, M, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-23 | | Release date: | 2009-07-07 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Northeast Structural Genomics Consortium Target TeR59
To be Published
|
|
4AO6
 
 | | Native structure of a novel cold-adapted esterase from an Arctic intertidal metagenomic library | | Descriptor: | ESTERASE | | Authors: | Fu, J, Leiros, H.-K.S, Pascale, D.d, Johnson, K.A, Blencke, H.M, Landfald, B. | | Deposit date: | 2012-03-23 | | Release date: | 2012-08-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Functional and Structural Studies of a Novel Cold-Adapted Esterase from an Arctic Intertidal Metagenomic Library.
Appl.Microbiol.Biotechnol., 97, 2013
|
|
4A7I
 
 | | Factor Xa in complex with a potent 2-amino-ethane sulfonamide inhibitor | | Descriptor: | 5-CHLORO-THIOPHENE-2-CARBOXYLIC ACID [2-(1--ISOPROPYL-PIPERIDIN-4-YLSULFAMOYL)-ETHYL]-AMIDE, ACTIVATED FACTOR XA HEAVY CHAIN XA, CALCIUM ION, ... | | Authors: | Nazare, M, Matter, H, Will, D.W, Wagner, M, Urmann, M, Czech, J, Schreuder, H, Bauer, A, Ritter, K, Wehner, V. | | Deposit date: | 2011-11-14 | | Release date: | 2012-02-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Fragment Deconstruction of Small, Potent Factor Xa Inhibitors: Exploring the Superadditivity Energetics of Fragment Linking in Protein-Ligand Complexes.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
4AN2
 
 | | Crystal structures of human MEK1 with carboxamide-based allosteric inhibitor XL518 (GDC-0973), or related analogs. | | Descriptor: | DUAL SPECIFICITY MITOGEN-ACTIVATED PROTEIN KINASE KINASE 1, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Rice, K.D, Aay, N, Anand, N.K, Blazey, C.M, Bowles, O.J, Bussenius, J, Costanzo, S, Curtis, J.K, Defina, S.C, Dubenko, L, Engst, S, Joshi, A.A, Kennedy, A.R, Kim, A.I, Koltun, E.S, Lougheed, J.C, Manalo, J.C.L, Martini, J.F, Nuss, J.M, Peto, C.J, Tsang, T.H, Yu, P, Johnston, S. | | Deposit date: | 2012-03-14 | | Release date: | 2012-12-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Novel Carboxamide-Based Allosteric Mek Inhibitors: Discovery and Optimization Efforts Toward Xl518 (Gdc-0973)
Acs Med.Chem.Lett., 3, 2012
|
|
4MI5
 
 | | Crystal structure of the EZH2 SET domain | | Descriptor: | Histone-lysine N-methyltransferase EZH2, SULFATE ION, ZINC ION | | Authors: | Antonysamy, S, Condon, B, Druzina, Z, Bonanno, J, Gheyi, T, Macewan, I, Zhang, A, Ashok, S, Russell, M, Luz, J.G. | | Deposit date: | 2013-08-30 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Context of Disease-Associated Mutations and Putative Mechanism of Autoinhibition Revealed by X-Ray Crystallographic Analysis of the EZH2-SET Domain.
Plos One, 8, 2013
|
|
4ANB
 
 | | Crystal structures of human MEK1 with carboxamide-based allosteric inhibitor XL518 (GDC-0973), or related analogs. | | Descriptor: | DUAL SPECIFICITY MITOGEN-ACTIVATED PROTEIN KINASE KINASE 1, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Rice, K.D, Aay, N, Anand, N.K, Blazey, C.M, Bowles, O.J, Bussenius, J, Costanzo, S, Curtis, J.K, Defina, S.C, Dubenko, L, Engst, S, Joshi, A.A, Kennedy, A.R, Kim, A.I, Koltun, E.S, Lougheed, J.C, Manalo, J.C.L, Martini, J.F, Nuss, J.M, Peto, C.J, Tsang, T.H, Yu, P, Johnston, S. | | Deposit date: | 2012-03-16 | | Release date: | 2012-12-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Novel Carboxamide-Based Allosteric Mek Inhibitors: Discovery and Optimization Efforts Toward Xl518 (Gdc-0973)
Acs Med.Chem.Lett., 3, 2012
|
|
4ANF
 
 | | Structure of the ornithine carbamoyltransferase from Mycoplasma penetrans with a P23 Space group | | Descriptor: | ORNITHINE CARBAMOYLTRANSFERASE, CATABOLIC | | Authors: | Gallego, P, Benach, J, Planell, R, Querol, E, Perez-Pons, J.A, Reverter, D. | | Deposit date: | 2012-03-16 | | Release date: | 2012-03-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Characterization of the Enzymes Composing the Arginine Deiminase Pathway in Mycoplasma Penetrans.
Plos One, 7, 2012
|
|
4AM0
 
 | | Structure of Dengue virus strain 4 DIII in complex with Fab 2H12 | | Descriptor: | ENVELOPE PROTEIN,, FAB 2H12, HEAVY CHAIN, ... | | Authors: | Midgley, C.M, Flanagan, A, Mongkolsapaya, J, Grimes, J.M, Screaton, G.R. | | Deposit date: | 2012-03-06 | | Release date: | 2012-06-06 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Structural Analysis of a Dengue Cross-Reactive Antibody Complexed with Envelope Domain III Reveals the Molecular Basis of Cross-Reactivity.
J.Immunol., 188, 2012
|
|
3HNR
 
 | | Crystal Structure of a probable methyltransferase BT9727_4108 from Bacillus thuringiensis subsp. Northeast Structural Genomics Consortium target id BuR219 | | Descriptor: | probable methyltransferase BT9727_4108 | | Authors: | Seetharaman, J, Neely, H, Sahdev, S, Janjua, H, Xiao, R, Ciccosanti, C, Foote, E.L, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-05-31 | | Release date: | 2009-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of a probable methyltransferase BT9727_4108 from Bacillus thuringiensis subsp. Northeast Structural Genomics Consortium target id BuR219
To be Published
|
|
3I3U
 
 | | Crystal structure of lp_1913 protein from lactobacillus plantarum, northeast structural genomics Consortium target lpr140a | | Descriptor: | uncharacterized protein lp_1913 | | Authors: | Seetharaman, J, Chen, Y, Forouhar, F, Sahdev, S, Janjua, H, Xiao, R, Ciccosanti, C, Foote, E.L, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-30 | | Release date: | 2009-07-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of lp_1913 protein from lactobacillus plantarum, northeast structural genomics Consortium target lpr140a
To be Published
|
|
4AMO
 
 | |
3HQ9
 
 | | CcpA from G. sulfurreducens, S134P variant | | Descriptor: | (R,R)-2,3-BUTANEDIOL, CALCIUM ION, Cytochrome c551 peroxidase, ... | | Authors: | Hoffmann, M, Seidel, J, Einsle, O. | | Deposit date: | 2009-06-05 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | CcpA from Geobacter sulfurreducens is a basic di-heme cytochrome c peroxidase.
J.Mol.Biol., 393, 2009
|
|
4AR2
 
 | | Dodecahedron formed of penton base protein from adenovirus Ad3 | | Descriptor: | CALCIUM ION, FIBER PROTEIN, L2 PROTEIN III (PENTON BASE) | | Authors: | Burmeister, W.P, Szolajska, E, Zochowska, M, Nerlo, B, Andreev, I, Schoehn, G, Andrieu, J.-P, Fender, P, Naskalska, A, Zubieta, C, Cusack, S, Chroboczek, J. | | Deposit date: | 2012-04-20 | | Release date: | 2012-10-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | The Structural Basis for the Integrity of Adenovirus Ad3 Dodecahedron.
Plos One, 7, 2012
|
|
4AN3
 
 | | Crystal structures of human MEK1 with carboxamide-based allosteric inhibitor XL518 (GDC-0973), or related analogs. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DUAL SPECIFICITY MITOGEN-ACTIVATED PROTEIN KINASE KINASE 1, MAGNESIUM ION, ... | | Authors: | Rice, K.D, Aay, N, Anand, N.K, Blazey, C.M, Bowles, O.J, Bussenius, J, Costanzo, S, Curtis, J.K, Defina, S.C, Dubenko, L, Engst, S, Joshi, A.A, Kennedy, A.R, Kim, A.I, Koltun, E.S, Lougheed, J.C, Manalo, J.C.L, Martini, J.F, Nuss, J.M, Peto, C.J, Tsang, T.H, Yu, P, Johnston, S. | | Deposit date: | 2012-03-15 | | Release date: | 2012-12-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel Carboxamide-Based Allosteric Mek Inhibitors: Discovery and Optimization Efforts Toward Xl518 (Gdc-0973)
Acs Med.Chem.Lett., 3, 2012
|
|
