3WW1
 
 | | X-ray structure of Cellulomonas parahominis L-ribose isomerase with L-ribose | | Descriptor: | L-ribose, L-ribose isomerase, MANGANESE (II) ION, ... | | Authors: | Terami, Y, Yoshida, H, Takata, G, Kamitori, S. | | Deposit date: | 2014-06-13 | | Release date: | 2015-04-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Essentiality of tetramer formation of Cellulomonas parahominis L-ribose isomerase involved in novel L-ribose metabolic pathway.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
3WT1
 
 | | Crystal structure of the b'-a' domain of thermophilic fungal protein disulfide isomerase (reduced form) | | Descriptor: | GLYCEROL, Protein disulfide-isomerase | | Authors: | Inagaki, K, Satoh, T, Itoh, S.G, Okumura, H, Kato, K. | | Deposit date: | 2014-04-02 | | Release date: | 2014-11-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Redox-dependent conformational transition of catalytic domain of protein disulfide isomerase indicated by crystal structure-based molecular dynamics simulation
Chem.Phys.Lett., 618, 2015
|
|
3W7U
 
 | | Escherichia coli K12 YgjK complexed with galactose | | Descriptor: | CALCIUM ION, Uncharacterized protein YgjK, alpha-D-galactopyranose | | Authors: | Miyazaki, T, Kurakata, Y, Uechi, A, Yoshida, H, Kamitori, S, Sakano, Y, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2013-03-06 | | Release date: | 2013-04-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural insights into the substrate specificity and function of Escherichia coli K12 YgjK, a glucosidase belonging to the glycoside hydrolase family 63.
J.Mol.Biol., 381, 2008
|
|
3WW4
 
 | | X-ray structures of Cellulomonas parahominis L-ribose isomerase with L-allose | | Descriptor: | L-allose, L-ribose isomerase, MANGANESE (II) ION, ... | | Authors: | Terami, Y, Yoshida, H, Takata, G, Kamitori, S. | | Deposit date: | 2014-06-13 | | Release date: | 2015-04-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Essentiality of tetramer formation of Cellulomonas parahominis L-ribose isomerase involved in novel L-ribose metabolic pathway.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
3WT2
 
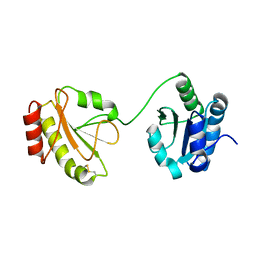 | | Crystal structure of the b'-a' domain of thermophilic fungal protein disulfide isomerase (oxidized form) | | Descriptor: | Protein disulfide-isomerase | | Authors: | Inagaki, K, Satoh, T, Itoh, S.G, Okumura, H, Kato, K. | | Deposit date: | 2014-04-02 | | Release date: | 2014-11-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Redox-dependent conformational transition of catalytic domain of protein disulfide isomerase indicated by crystal structure-based molecular dynamics simulation
Chem.Phys.Lett., 618, 2015
|
|
3WW2
 
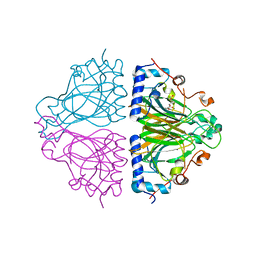 | | X-ray structures of Cellulomonas parahominis L-ribose isomerase with L-psicose | | Descriptor: | L-psicose, L-ribose isomerase, MANGANESE (II) ION, ... | | Authors: | Terami, Y, Yoshida, H, Takata, G, Kamitori, S. | | Deposit date: | 2014-06-13 | | Release date: | 2015-04-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Essentiality of tetramer formation of Cellulomonas parahominis L-ribose isomerase involved in novel L-ribose metabolic pathway.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
5WSF
 
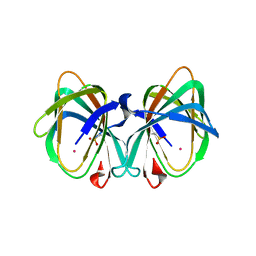 | | Crystal structure of a cupin protein (tm1459) in osmium (Os)-substituted form II | | Descriptor: | OSMIUM ION, Uncharacterized protein tm1459 | | Authors: | Fujieda, N, Nakano, T, Taniguchi, Y, Ichihashi, H, Nishikawa, Y, Kurisu, G, Itoh, S. | | Deposit date: | 2016-12-06 | | Release date: | 2017-05-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | A Well-Defined Osmium-Cupin Complex: Hyperstable Artificial Osmium Peroxygenase
J. Am. Chem. Soc., 2017
|
|
2D0H
 
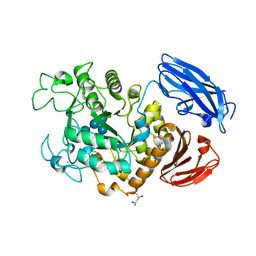 | | Crystal Structure of Thermoactinomyces vulgaris R-47 Alpha-Amylase 1 (TVAI) Mutant D356N/E396Q complexed with P2, a pullulan model oligosaccharide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Abe, A, Yoshida, H, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2005-08-02 | | Release date: | 2006-07-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Complexes of Thermoactinomyces vulgaris R-47 alpha-amylase 1 and pullulan model oligossacharides provide new insight into the mechanism for recognizing substrates with alpha-(1,6) glycosidic linkages
Febs J., 272, 2005
|
|
5X2J
 
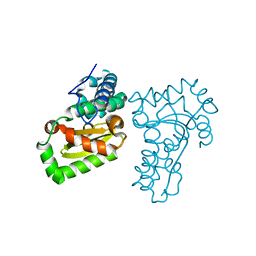 | | Crystal structure of a recombinant hybrid manganese superoxide dismutase from Staphylococcus equorum and Staphylococcus saprophyticus | | Descriptor: | MANGANESE (II) ION, manganese superoxide dismutase | | Authors: | Retnoningrum, D.S, Yoshida, H, Arumsari, S, Kamitori, S, Ismaya, W.T. | | Deposit date: | 2017-01-31 | | Release date: | 2018-01-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The first crystal structure of manganese superoxide dismutase from the genus Staphylococcus
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
2D0F
 
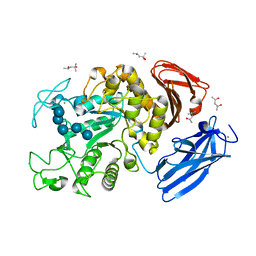 | | Crystal Structure of Thermoactinomyces vulgaris R-47 Alpha-Amylase 1 (TVAI) Mutant D356N complexed with P2, a pullulan model oligosaccharide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Abe, A, Yoshida, H, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2005-08-02 | | Release date: | 2006-07-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Complexes of Thermoactinomyces vulgaris R-47 alpha-amylase 1 and pullulan model oligossacharides provide new insight into the mechanism for recognizing substrates with alpha-(1,6) glycosidic linkages
Febs J., 272, 2005
|
|
5WSD
 
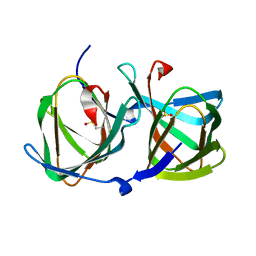 | | Crystal structure of a cupin protein (tm1459) in apo form | | Descriptor: | Uncharacterized protein tm1459 | | Authors: | Fujieda, N, Nakano, T, Taniguchi, Y, Ichihashi, H, Nishikawa, Y, Kurisu, G, Itoh, S. | | Deposit date: | 2016-12-06 | | Release date: | 2017-05-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | A Well-Defined Osmium-Cupin Complex: Hyperstable Artificial Osmium Peroxygenase
J. Am. Chem. Soc., 2017
|
|
5WSE
 
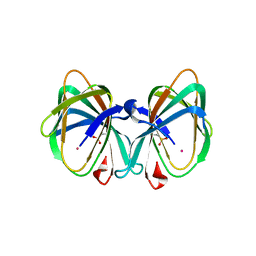 | | Crystal structure of a cupin protein (tm1459) in osmium (Os) substituted form I | | Descriptor: | OSMIUM ION, Uncharacterized protein tm1459 | | Authors: | Fujieda, N, Nakano, T, Taniguchi, Y, Ichihashi, H, Nishikawa, Y, Kurisu, G, Itoh, S. | | Deposit date: | 2016-12-06 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | A Well-Defined Osmium-Cupin Complex: Hyperstable Artificial Osmium Peroxygenase
J. Am. Chem. Soc., 2017
|
|
3ITY
 
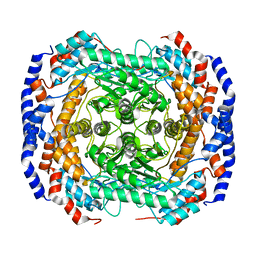 | | Metal-free form of Pseudomonas stutzeri L-rhamnose isomerase | | Descriptor: | L-rhamnose isomerase | | Authors: | Yoshida, H, Yamaji, M, Ishii, T, Izumori, K, Kamitori, S. | | Deposit date: | 2009-08-28 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Catalytic reaction mechanism of Pseudomonas stutzeri l-rhamnose isomerase deduced from X-ray structures
Febs J., 277, 2010
|
|
8F2R
 
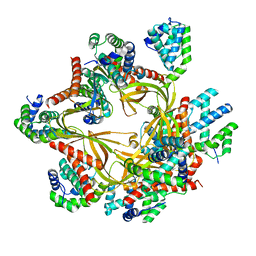 | | Human CCC complex | | Descriptor: | COMM domain-containing protein 1, COMM domain-containing protein 10, COMM domain-containing protein 2, ... | | Authors: | Healy, M.D, McNally, K.E, Butkovic, R, Chilton, M, Kato, K, Sacharz, J, McConville, C, Moody, E.R.R, Shaw, S, Planelles-Herrero, V.J, Kadapalakere, S.Y, Ross, J, Borucu, U, Palmer, C.S, Chen, K, Croll, T.I, Hall, R.J, Caruana, N.J, Ghai, R, Nguyen, T.H.D, Heesom, K.J, Saitoh, S, Berger, I, Berger-Schaffitzel, C, Williams, T.A, Stroud, D.A, Derivery, E, Collins, B.M, Cullen, P.J. | | Deposit date: | 2022-11-08 | | Release date: | 2023-05-24 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Structure of the endosomal Commander complex linked to Ritscher-Schinzel syndrome.
Cell, 186, 2023
|
|
8F2U
 
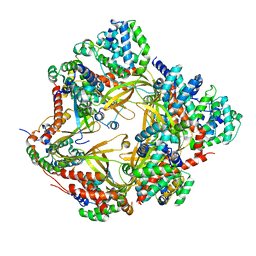 | | Human CCC complex | | Descriptor: | COMM domain-containing protein 1, COMM domain-containing protein 10, COMM domain-containing protein 2, ... | | Authors: | Healy, M.D, McNally, K.E, Butkovic, R, Chilton, M, Kato, K, Sacharz, J, McConville, C, Moody, E.R.R, Shaw, S, Planelles-Herrero, V.J, Kadapalakere, S.Y, Ross, J, Borucu, U, Palmer, C.S, Chen, K, Croll, T.I, Hall, R.J, Caruana, N.J, Ghai, R, Nguyen, T.H.D, Heesom, K.J, Saitoh, S, Berger, I, Berger-Schaffitzel, C, Williams, T.A, Stroud, D.A, Derivery, E, Collins, B.M, Cullen, P.J. | | Deposit date: | 2022-11-08 | | Release date: | 2023-05-24 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Structure of the endosomal Commander complex linked to Ritscher-Schinzel syndrome.
Cell, 186, 2023
|
|
3IUD
 
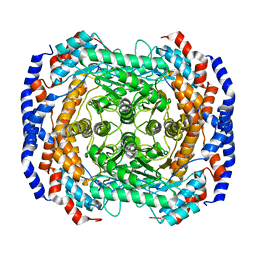 | | Cu2+-bound form of Pseudomonas stutzeri L-rhamnose isomerase | | Descriptor: | COPPER (II) ION, L-rhamnose isomerase | | Authors: | Yoshida, H, Yamaji, M, Ishii, T, Izumori, K, Kamitori, S. | | Deposit date: | 2009-08-31 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Catalytic reaction mechanism of Pseudomonas stutzeri l-rhamnose isomerase deduced from X-ray structures
Febs J., 277, 2010
|
|
3IUI
 
 | | Zn2+-bound form of Pseudomonas stutzeri L-rhamnose isomerase | | Descriptor: | L-rhamnose isomerase, ZINC ION | | Authors: | Yoshida, H, Yamaji, M, Ishii, T, Izumori, K, Kamitori, S. | | Deposit date: | 2009-08-31 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Catalytic reaction mechanism of Pseudomonas stutzeri l-rhamnose isomerase deduced from X-ray structures
Febs J., 277, 2010
|
|
3ITX
 
 | | Mn2+ bound form of Pseudomonas stutzeri L-rhamnose isomerase | | Descriptor: | L-rhamnose isomerase, MANGANESE (II) ION | | Authors: | Yoshida, H, Yamaji, M, Ishii, T, Izumori, K, Kamitori, S. | | Deposit date: | 2009-08-28 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Catalytic reaction mechanism of Pseudomonas stutzeri l-rhamnose isomerase deduced from X-ray structures
Febs J., 277, 2010
|
|
2D0G
 
 | | Crystal Structure of Thermoactinomyces vulgaris R-47 Alpha-Amylase 1 (TVAI) Mutant D356N/E396Q complexed with P5, a pullulan model oligosaccharide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, alpha-D-glucopyranose, ... | | Authors: | Abe, A, Yoshida, H, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2005-08-02 | | Release date: | 2006-07-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Complexes of Thermoactinomyces vulgaris R-47 alpha-amylase 1 and pullulan model oligossacharides provide new insight into the mechanism for recognizing substrates with alpha-(1,6) glycosidic linkages
Febs J., 272, 2005
|
|
3IUH
 
 | | Co2+-bound form of Pseudomonas stutzeri L-rhamnose isomerase | | Descriptor: | COBALT (II) ION, L-rhamnose isomerase | | Authors: | Yoshida, H, Yamaji, M, Ishii, T, Izumori, K, Kamitori, S. | | Deposit date: | 2009-08-31 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Catalytic reaction mechanism of Pseudomonas stutzeri l-rhamnose isomerase deduced from X-ray structures
Febs J., 277, 2010
|
|
5J8L
 
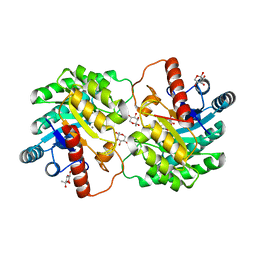 | | Crystal structure of D-tagatose 3-epimerase C66S from Pseudomonas cichorii in complex with 1-deoxy L-tagatose, using a crystal grown in microgravity | | Descriptor: | 1-deoxy-L-tagatose, 1-deoxy-beta-L-tagatopyranose, D-tagatose 3-epimerase, ... | | Authors: | Yoshida, H, Yoshihara, A, Izumori, K, Kamitori, S. | | Deposit date: | 2016-04-08 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | X-ray structures of the Pseudomonas cichorii D-tagatose 3-epimerase mutant form C66S recognizing deoxy sugars as substrates
Appl. Microbiol. Biotechnol., 100, 2016
|
|
5ZFS
 
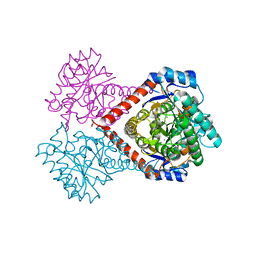 | | Crystal structure of Arthrobacter globiformis M30 sugar epimerase which can produce D-allulose from D-fructose | | Descriptor: | ACETATE ION, D-allulose-3-epimerase, MANGANESE (II) ION | | Authors: | Yoshida, H, Yoshihara, A, Gullapalli, P.K, Ohtani, K, Akimitsu, K, Izumori, K, Kamitori, S. | | Deposit date: | 2018-03-07 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | X-ray structure of Arthrobacter globiformis M30 ketose 3-epimerase for the production of D-allulose from D-fructose.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
4H2R
 
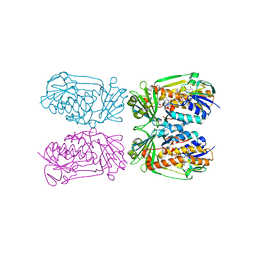 | | Structure of MHPCO Y270F mutant, 5-hydroxynicotinic acid complex | | Descriptor: | 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, 5-hydroxypyridine-3-carboxylic acid, BETA-MERCAPTOETHANOL, ... | | Authors: | Kobayashi, J, Yoshida, H, Mikami, B, Hayashi, H, Kamitori, S, Yagi, T. | | Deposit date: | 2012-09-13 | | Release date: | 2013-09-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.473 Å) | | Cite: | Crystal structure of 2-Methyl-3-hydroxypyridine-5-carboxylic acid oxygenase
To be Published
|
|
7X7O
 
 | | SARS-CoV-2 spike RBD in complex with neutralizing antibody UT28K | | Descriptor: | Spike protein S1, UT28K Fab, heavy chain, ... | | Authors: | Ozawa, T, Tani, H, Anraku, Y, Kita, S, Igarashi, E, Saga, Y, Inasaki, N, Kawasuji, H, Yamada, H, Sasaki, S, Somekawa, M, Sasaki, J, Hayakawa, Y, Yamamoto, Y, Morinaga, Y, Kurosawa, N, Isobe, M, Fukuhara, H, Maenaka, K, Hashiguchi, T, Kishi, H, Kitajima, I, Saito, S, Niimi, H. | | Deposit date: | 2022-03-10 | | Release date: | 2022-05-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | Novel super-neutralizing antibody UT28K is capable of protecting against infection from a wide variety of SARS-CoV-2 variants.
Mabs, 14, 2022
|
|
2D2O
 
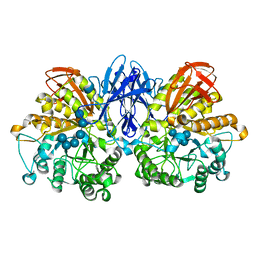 | | Structure of a complex of Thermoactinomyces vulgaris R-47 alpha-amylase 2 with maltohexaose demonstrates the important role of aromatic residues at the reducing end of the substrate binding cleft | | Descriptor: | CALCIUM ION, Neopullulanase 2, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Ohtaki, A, Mizuno, M, Yoshida, H, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2005-09-13 | | Release date: | 2006-08-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a complex of Thermoactinomyces vulgaris R-47 alpha-amylase 2 with maltohexaose demonstrates the important role of aromatic residues at the reducing end of the substrate binding cleft
Carbohydr.Res., 341, 2006
|
|
