1WN7
 
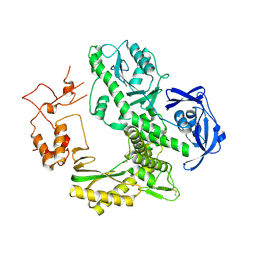 | | Crystal structure of archaeal family B DNA polymerase mutant | | Descriptor: | Family B DNA Polymerase, GLYCEROL, NICKEL (II) ION | | Authors: | Kuroita, T, Matsumura, H, Yokota, N, Hashimoto, H, Imanaka, T, Inoue, T, Kai, Y. | | Deposit date: | 2004-07-28 | | Release date: | 2005-08-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural Mechanism for Coordination of Proofreading and Polymerase Activities in Archaeal DNA Polymerases
J.Mol.Biol., 351, 2005
|
|
7DF2
 
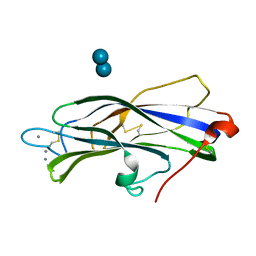 | |
5ZM9
 
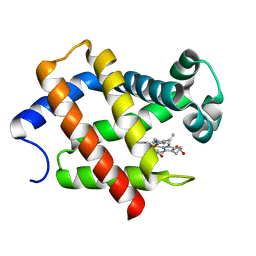 | | Crystal structure of hexacoordinated heme protein from anhydrobiotic tardigrade at pH 7 | | Descriptor: | CHLORIDE ION, Globin Protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kim, J, Fukuda, Y, Inoue, T. | | Deposit date: | 2018-04-02 | | Release date: | 2019-01-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of Kumaglobin: a hexacoordinated heme protein from an anhydrobiotic tardigrade, Ramazzottius varieornatus.
FEBS J., 286, 2019
|
|
5Z4G
 
 | |
5X31
 
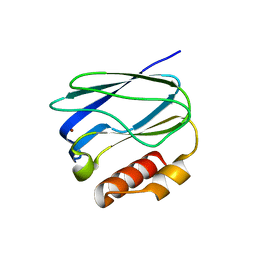 | |
5XDU
 
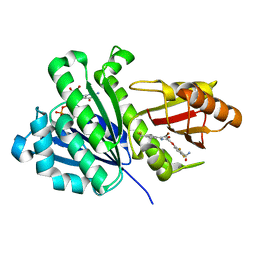 | | Staphylococcus aureus FtsZ 12-316 complexed with TXA6101 | | Descriptor: | 3-[[5-bromanyl-4-[4-(trifluoromethyl)phenyl]-1,3-oxazol-2-yl]methoxy]-2,6-bis(fluoranyl)benzamide, CALCIUM ION, Cell division protein FtsZ, ... | | Authors: | Fujita, J, Maeda, Y, Mizohata, E, Inoue, T, Kaul, M, Parhi, A.K, LaVoie, E.J, Pilch, D.S, Matsumura, H. | | Deposit date: | 2017-03-30 | | Release date: | 2017-08-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Flexibility of an Inhibitor Overcomes Drug Resistance Mutations in Staphylococcus aureus FtsZ
ACS Chem. Biol., 12, 2017
|
|
5XDV
 
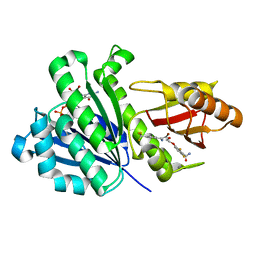 | | Staphylococcus aureus FtsZ 12-316 G196S complexed with TXA6101 | | Descriptor: | 3-[[5-bromanyl-4-[4-(trifluoromethyl)phenyl]-1,3-oxazol-2-yl]methoxy]-2,6-bis(fluoranyl)benzamide, CALCIUM ION, Cell division protein FtsZ, ... | | Authors: | Fujita, J, Maeda, Y, Mizohata, E, Inoue, T, Kaul, M, Parhi, A.K, LaVoie, E.J, Pilch, D.S, Matsumura, H. | | Deposit date: | 2017-03-30 | | Release date: | 2017-08-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Flexibility of an Inhibitor Overcomes Drug Resistance Mutations in Staphylococcus aureus FtsZ
ACS Chem. Biol., 12, 2017
|
|
5XDT
 
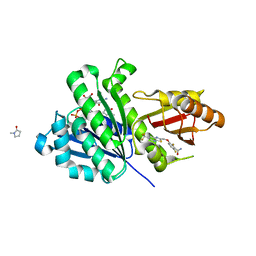 | | Staphylococcus aureus FtsZ 12-316 complexed with TXA707 | | Descriptor: | 1-methylpyrrolidin-2-one, 2,6-bis(fluoranyl)-3-[[6-(trifluoromethyl)-[1,3]thiazolo[5,4-b]pyridin-2-yl]methoxy]benzamide, CALCIUM ION, ... | | Authors: | Fujita, J, Maeda, Y, Mizohata, E, Inoue, T, Kaul, M, Parhi, A.K, LaVoie, E.J, Pilch, D.S, Matsumura, H. | | Deposit date: | 2017-03-30 | | Release date: | 2017-08-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural Flexibility of an Inhibitor Overcomes Drug Resistance Mutations in Staphylococcus aureus FtsZ
ACS Chem. Biol., 12, 2017
|
|
5XDW
 
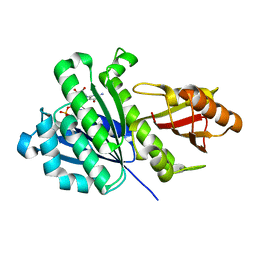 | | Staphylococcus aureus FtsZ 12-316 G196S | | Descriptor: | CALCIUM ION, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Fujita, J, Maeda, Y, Mizohata, E, Inoue, T, Kaul, M, Parhi, A.K, LaVoie, E.J, Pilch, D.S, Matsumura, H. | | Deposit date: | 2017-03-30 | | Release date: | 2017-08-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Flexibility of an Inhibitor Overcomes Drug Resistance Mutations in Staphylococcus aureus FtsZ
ACS Chem. Biol., 12, 2017
|
|
1WDD
 
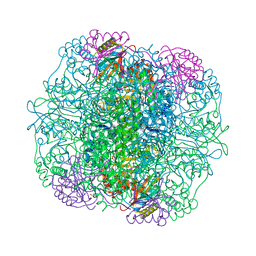 | | Crystal Structure of Activated Rice Rubisco Complexed with 2-Carboxyarabinitol-1,5-bisphosphate | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Mizohata, E, Matsumura, H, Ueno, T, Ishida, H, Inoue, T, Makino, A, Mae, T, Kai, Y. | | Deposit date: | 2004-05-13 | | Release date: | 2004-11-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of rice Rubisco and implications for activation induced by positive effectors NADPH and 6-phosphogluconate
J.Mol.Biol., 422, 2012
|
|
1WM1
 
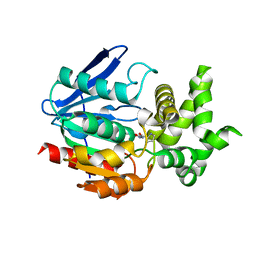 | | Crystal Structure of Prolyl Aminopeptidase, Complex with Pro-TBODA | | Descriptor: | (5-TERT-BUTYL-1,3,4-OXADIAZOL-2-YL)[(2R)-PYRROLIDIN-2-YL]METHANONE, Proline iminopeptidase | | Authors: | Nakajima, Y, Inoue, T, Ito, K, Tozaka, T, Hatakeyama, S, Tanaka, N, Nakamura, K.T, Yoshimoto, T. | | Deposit date: | 2004-07-01 | | Release date: | 2004-07-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel inhibitor for prolyl aminopeptidase from Serratia marcescens and studies on the mechanism of substrate recognition of the enzyme using the inhibitor
ARCH.BIOCHEM.BIOPHYS., 416, 2003
|
|
1X0R
 
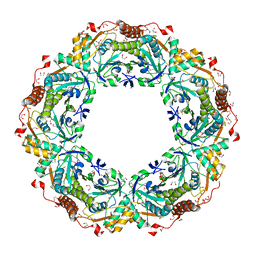 | | Thioredoxin Peroxidase from Aeropyrum pernix K1 | | Descriptor: | 1,2-ETHANEDIOL, Probable peroxiredoxin | | Authors: | Nakamura, T, Yamamoto, T, Inoue, T, Matsumura, H, Kobayashi, A, Hagihara, Y, Uegaki, K, Ataka, M, Kai, Y, Ishikawa, K. | | Deposit date: | 2005-03-28 | | Release date: | 2005-12-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of thioredoxin peroxidase from aerobic hyperthermophilic archaeon Aeropyrum pernix K1
Proteins, 62, 2006
|
|
1WLJ
 
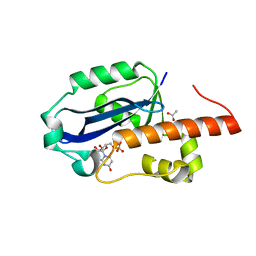 | | human ISG20 | | Descriptor: | ACETATE ION, MANGANESE (II) ION, URIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Horio, T, Murai, M, Inoue, T, Hamasaki, T, Tanaka, T, Ohgi, T. | | Deposit date: | 2004-06-28 | | Release date: | 2004-12-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human ISG20, an interferon-induced antiviral ribonuclease
Febs Lett., 577, 2004
|
|
1WRF
 
 | | Refined solution structure of Der f 2, The Major Mite Allergen from Dermatophagoides farinae | | Descriptor: | Mite group 2 allergen Der f 2 | | Authors: | Ichikawa, S, Takai, T, Inoue, T, Yuuki, T, Okumura, Y, Ogura, K, Inagaki, F, Hatanaka, H. | | Deposit date: | 2004-10-15 | | Release date: | 2005-04-19 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | NMR Study on the Major Mite Allergen Der f 2: Its Refined Tertiary Structure, Epitopes for Monoclonal Antibodies and Characteristics Shared by ML Protein Group Members
J.Biochem.(Tokyo), 137, 2005
|
|
1WNS
 
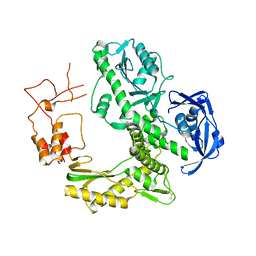 | | Crystal structure of family B DNA polymerase from hyperthermophilic archaeon pyrococcus kodakaraensis KOD1 | | Descriptor: | DNA POLYMERASE | | Authors: | Hashimoto, H, Inoue, T, Kai, Y, Fujiwara, S, Takagi, M, Nishioka, M, Imanaka, T. | | Deposit date: | 2004-08-09 | | Release date: | 2004-08-17 | | Last modified: | 2017-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of DNA Polymerase from Hyperthermophilic Archaeon Pyrococcus Kodakaraensis Kod1
J.Mol.Biol., 306, 2001
|
|
6J27
 
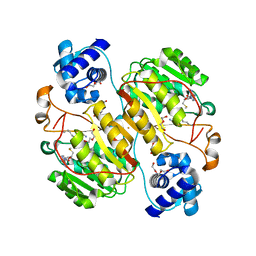 | | Crystal structure of the branched-chain polyamine synthase from Thermus thermophilus (Tth-BpsA) in complex with N4-aminopropylspermidine and 5'-methylthioadenosine | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, DI(HYDROXYETHYL)ETHER, FE (III) ION, ... | | Authors: | Mizohata, E, Toyoda, M, Fujita, J, Inoue, T. | | Deposit date: | 2018-12-31 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | The C-terminal flexible region of branched-chain polyamine synthase facilitates substrate specificity and catalysis.
Febs J., 286, 2019
|
|
6J28
 
 | | Crystal structure of the branched-chain polyamine synthase C9 mutein from Thermus thermophilus (Tth-BpsA C9) in complex with N4-aminopropylspermidine and 5'-methylthioadenosine | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5'-DEOXY-5'-METHYLTHIOADENOSINE, FE (III) ION, ... | | Authors: | Mizohata, E, Toyoda, M, Fujita, J, Inoue, T. | | Deposit date: | 2018-12-31 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The C-terminal flexible region of branched-chain polyamine synthase facilitates substrate specificity and catalysis.
Febs J., 286, 2019
|
|
6J26
 
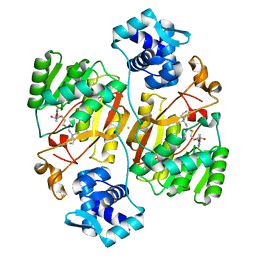 | | Crystal structure of the branched-chain polyamine synthase from Thermococcus kodakarensis (Tk-BpsA) in complex with N4-bis(aminopropyl)spermidine and 5'-methylthioadenosine | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, FE (III) ION, N(4)-bis(aminopropyl)spermidine synthase, ... | | Authors: | Mizohata, E, Toyoda, M, Fujita, J, Inoue, T. | | Deposit date: | 2018-12-31 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The C-terminal flexible region of branched-chain polyamine synthase facilitates substrate specificity and catalysis.
Febs J., 286, 2019
|
|
1WS8
 
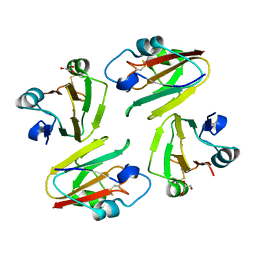 | | Crystal Structure of Mavicyanin from Cucurbita pepo medullosa (Zucchini) | | Descriptor: | COPPER (II) ION, GLYCEROL, mavicyanin | | Authors: | Xie, Y, Inoue, T, Miyamoto, Y, Matsumura, H, Kunishige, K, Yamaguchi, K, Nojini, M, Suzuki, S, Kai, Y. | | Deposit date: | 2004-11-02 | | Release date: | 2004-11-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural reorganization of the copper binding site involving Thr15 of mavicyanin from Cucurbita pepo medullosa (zucchini) upon reduction.
J.Biochem.(Tokyo), 137, 2005
|
|
1WS7
 
 | | Crystal Structure of Mavicyanin from Cucurbita pepo medullosa (Zucchini) | | Descriptor: | COPPER (I) ION, Mavicyanin | | Authors: | Xie, Y, Inoue, T, Miyamoto, Y, Matsumura, H, Kataoka, K, Yamaguchi, K, Nojini, M, Suzuki, S, Kai, Y. | | Deposit date: | 2004-11-02 | | Release date: | 2004-11-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural reorganization of the copper binding site involving Thr15 of mavicyanin from Cucurbita pepo medullosa (zucchini) upon reduction.
J.Biochem.(Tokyo), 137, 2005
|
|
1RKR
 
 | | CRYSTAL STRUCTURE OF AZURIN-I FROM ALCALIGENES XYLOSOXIDANS NCIMB 11015 | | Descriptor: | AZURIN-I, COPPER (II) ION | | Authors: | Li, C, Inoue, T, Gotowda, M, Suzuki, S, Yamaguchi, K, Kataoka, K, Kai, Y. | | Deposit date: | 1997-05-17 | | Release date: | 1998-05-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of azurin I from the denitrifying bacterium Alcaligenes xylosoxidans NCIMB 11015 at 2.45 A resolution.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1V7Z
 
 | | creatininase-product complex | | Descriptor: | MANGANESE (II) ION, N-[(E)-AMINO(IMINO)METHYL]-N-METHYLGLYCINE, SULFATE ION, ... | | Authors: | Yoshimoto, T, Tanaka, N, Kanada, N, Inoue, T, Nakajima, Y, Haratake, M, Nakamura, K.T, Xu, Y, Ito, K. | | Deposit date: | 2003-12-26 | | Release date: | 2004-01-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of creatininase reveal the substrate binding site and provide an insight into the catalytic mechanism
J.Mol.Biol., 337, 2004
|
|
2Z5G
 
 | | Crystal structure of T1 lipase F16L mutant | | Descriptor: | CALCIUM ION, CHLORIDE ION, Thermostable lipase, ... | | Authors: | Matsumura, H, Yamamoto, T, Inoue, T, Kai, Y. | | Deposit date: | 2007-07-08 | | Release date: | 2007-10-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Novel cation-pi interaction revealed by crystal structure of thermoalkalophilic lipase
Proteins, 70, 2007
|
|
3A5W
 
 | | Peroxiredoxin (wild type) from Aeropyrum pernix K1 (reduced form) | | Descriptor: | Probable peroxiredoxin | | Authors: | Nakamura, T, Kado, Y, Yamaguchi, T, Matsumura, H, Ishikawa, K, Inoue, T. | | Deposit date: | 2009-08-12 | | Release date: | 2009-10-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of peroxiredoxin from Aeropyrum pernix K1 complexed with its substrate, hydrogen peroxide
J.Biochem., 147, 2010
|
|
2ZCT
 
 | | Oxidation of archaeal peroxiredoxin involves a hypervalent sulfur intermediate | | Descriptor: | Probable peroxiredoxin | | Authors: | Nakamura, T, Hagihara, Y, Abe, M, Inoue, T, Yamamoto, T, Matsumura, H. | | Deposit date: | 2007-11-12 | | Release date: | 2008-05-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Oxidation of archaeal peroxiredoxin involves a hypervalent sulfur intermediate
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
