1WM5
 
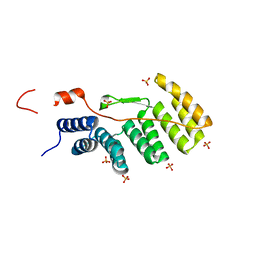 | |
1EPH
 
 | |
3VH4
 
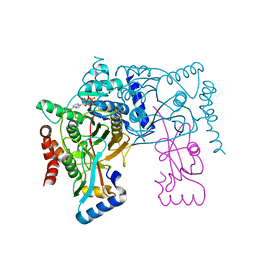 | | Crystal structure of Atg7CTD-Atg8-MgATP complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Autophagy-related protein 8, MAGNESIUM ION, ... | | Authors: | Noda, N.N, Satoo, K, Inagaki, F. | | Deposit date: | 2011-08-23 | | Release date: | 2011-09-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis of Atg8 activation by a homodimeric E1, Atg7.
Mol.Cell, 44, 2011
|
|
3VU0
 
 | | Crystal structure of the C-terminal globular domain of oligosaccharyltransferase (AfAglB-S2, AF_0040, O30195_ARCFU) from Archaeoglobus fulgidus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Putative uncharacterized protein | | Authors: | Nyirenda, J, Matsumoto, S, Saitoh, T, Maita, N, Noda, N.N, Inagaki, F, Kohda, D. | | Deposit date: | 2012-06-13 | | Release date: | 2013-01-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystallographic and NMR Evidence for Flexibility in Oligosaccharyltransferases and Its Catalytic Significance
Structure, 21, 2013
|
|
3VP7
 
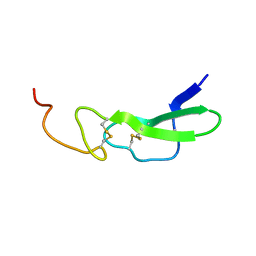
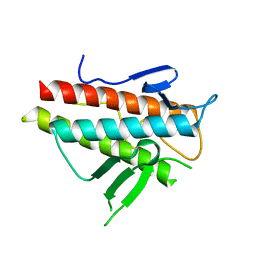 | | Crystal structure of the beta-alpha repeated, autophagy-specific (BARA) domain of Vps30/Atg6 | | Descriptor: | Vacuolar protein sorting-associated protein 30 | | Authors: | Noda, N.N, Kobayashi, T, Adachi, W, Fujioka, Y, Ohsumi, Y, Inagaki, F. | | Deposit date: | 2012-02-28 | | Release date: | 2012-03-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the novel C-terminal domain of vacuolar protein sorting 30/autophagy-related protein 6 and its specific role in autophagy.
J.Biol.Chem., 287, 2012
|
|
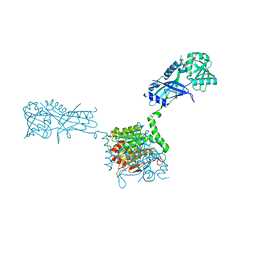
3VH2
 
 | |
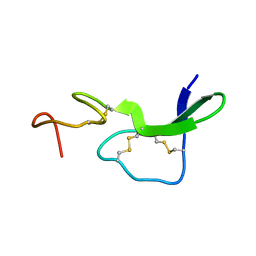
1EPI
 
 | |
2RQV
 
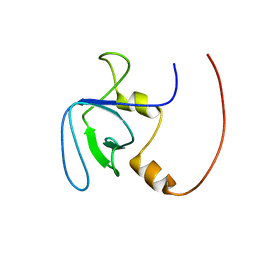 | |
2RQW
 
 | |
2RPV
 
 | | Solution Structure of GB1 with LBT probe | | Descriptor: | Immunoglobulin G-binding protein G, LANTHANUM (III) ION | | Authors: | Saio, T, Ogura, K, Yokochi, M, Kobashigawa, Y, Inagaki, F. | | Deposit date: | 2008-10-28 | | Release date: | 2009-09-15 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Two-point anchoring of a lanthanide-binding peptide to a target protein enhances the paramagnetic anisotropic effect
J.Biomol.Nmr, 44, 2009
|
|
2RQE
 
 | | Solution structure of the silkworm bGRP/GNBP3 N-terminal domain reveals the mechanism for b-1,3-glucan specific recognition | | Descriptor: | Beta-1,3-glucan-binding protein | | Authors: | Takahasi, K, Ochiai, M, Horiuchi, M, Kumeta, H, Ogura, K, Ashida, M, Inagaki, F. | | Deposit date: | 2009-04-22 | | Release date: | 2009-06-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the silkworm betaGRP/GNBP3 N-terminal domain reveals the mechanism for beta-1,3-glucan-specific recognition.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1EPG
 
 | |
1EPJ
 
 | |
5H9V
 
 | | Crystal structure of Regnase PIN domain, form I | | Descriptor: | Ribonuclease ZC3H12A, SODIUM ION | | Authors: | Yokogawa, M, Tsushima, T, Adachi, W, Noda, N.N, Inagaki, F. | | Deposit date: | 2015-12-29 | | Release date: | 2016-03-16 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for the regulation of enzymatic activity of Regnase-1 by domain-domain interactions
Sci Rep, 6, 2016
|
|
5H9W
 
 | | Crystal structure of Regnase PIN domain, form II | | Descriptor: | Ribonuclease ZC3H12A, SODIUM ION | | Authors: | Yokogawa, M, Tsushima, T, Adachi, W, Noda, N.N, Inagaki, F. | | Deposit date: | 2015-12-29 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the regulation of enzymatic activity of Regnase-1 by domain-domain interactions
Sci Rep, 6, 2016
|
|
3VH3
 
 | | Crystal structure of Atg7CTD-Atg8 complex | | Descriptor: | Autophagy-related protein 8, Ubiquitin-like modifier-activating enzyme ATG7, ZINC ION | | Authors: | Noda, N.N, Satoo, K, Inagaki, F. | | Deposit date: | 2011-08-23 | | Release date: | 2011-09-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of Atg8 activation by a homodimeric E1, Atg7.
Mol.Cell, 44, 2011
|
|
3VGP
 
 | | Crystal structure of the C-terminal globular domain of oligosaccharyltransferase (AF_0329) from Archaeoglobus fulgidus | | Descriptor: | Transmembrane oligosaccharyl transferase, putative | | Authors: | Matsumoto, S, Igura, M, Nyirenda, J, Yuzawa, S, Noda, N.N, Inagaki, F, Kohda, D. | | Deposit date: | 2011-08-18 | | Release date: | 2012-07-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of the C-Terminal Globular Domain of Oligosaccharyltransferase from Archaeoglobus fulgidus at 1.75 A Resolution
Biochemistry, 51, 2012
|
|
4DVY
 
 | | Crystal structure of the Helicobacter pylori CagA oncoprotein | | Descriptor: | Cytotoxicity-associated immunodominant antigen | | Authors: | Hayashi, T, Senda, M, Morohashi, H, Higashi, H, Horio, M, Kashiba, Y, Nagase, L, Sasaya, D, Shimizu, T, Venugopalan, N, Kumeta, H, Noda, N, Inagaki, F, Senda, T, Hatakeyama, M. | | Deposit date: | 2012-02-23 | | Release date: | 2012-07-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Tertiary Structure-Function Analysis Reveals the Pathogenic Signaling Potentiation Mechanism of Helicobacter pylori Oncogenic Effector CagA
Cell Host Microbe, 12, 2012
|
|
4DVZ
 
 | | Crystal structure of the Helicobacter pylori CagA oncoprotein | | Descriptor: | Cytotoxicity-associated immunodominant antigen | | Authors: | Hayashi, T, Senda, M, Morohashi, H, Higashi, H, Horio, M, Kashiba, Y, Nagase, L, Sasaya, D, Shimizu, T, Venugopalan, N, Kumeta, H, Noda, N, Inagaki, F, Senda, T, Hatakeyama, M. | | Deposit date: | 2012-02-23 | | Release date: | 2012-07-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Tertiary structure-function analysis reveals the pathogenic signaling potentiation mechanism of Helicobacter pylori oncogenic effector CagA
Cell Host Microbe, 12, 2012
|
|
5JH9
 
 | | Crystal structure of prApe1 | | Descriptor: | CACODYLATE ION, Vacuolar aminopeptidase 1, ZINC ION | | Authors: | Noda, N.N, Adachi, W, Inagaki, F. | | Deposit date: | 2016-04-20 | | Release date: | 2016-06-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Receptor-Mediated Selective Autophagy of Aminopeptidase I Aggregates
Cell Rep, 16, 2016
|
|
5JGF
 
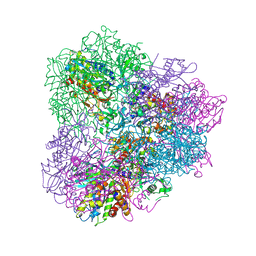 | | Crystal structure of mApe1 | | Descriptor: | Vacuolar aminopeptidase 1, ZINC ION | | Authors: | Noda, N.N, Adachi, W, Inagaki, F. | | Deposit date: | 2016-04-20 | | Release date: | 2016-06-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural Basis for Receptor-Mediated Selective Autophagy of Aminopeptidase I Aggregates
Cell Rep, 16, 2016
|
|
1IO6
 
 | |
1HSQ
 
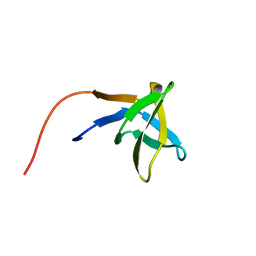 | | SOLUTION STRUCTURE OF THE SH3 DOMAIN OF PHOSPHOLIPASE CGAMMA | | Descriptor: | PHOSPHOLIPASE C-GAMMA (SH3 DOMAIN) | | Authors: | Kohda, D, Hatanaka, H, Odaka, M, Inagaki, F. | | Deposit date: | 1994-06-13 | | Release date: | 1994-08-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SH3 domain of phospholipase C-gamma.
Cell(Cambridge,Mass.), 72, 1993
|
|
1J2F
 
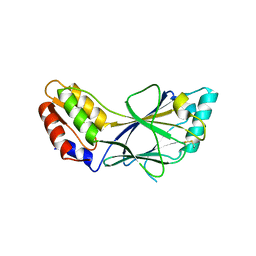 | | X-ray crystal structure of IRF-3 and its functional implications | | Descriptor: | Interferon regulatory factor 3 | | Authors: | Takahasi, K, Noda, N, Horiuchi, M, Mori, M, Okabe, Y, Fukuhara, Y, Terasawa, H, Fujita, T, Inagaki, F. | | Deposit date: | 2003-01-04 | | Release date: | 2003-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray crystal structure of IRF-3 and its functional implications.
Nat.Struct.Biol., 10, 2003
|
|
1L4V
 
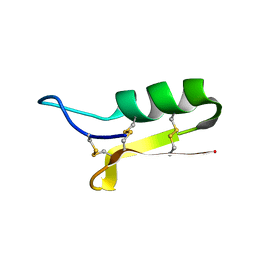 | | SOLUTION STRUCTURE OF SAPECIN | | Descriptor: | Sapecin | | Authors: | Hanzawa, H, Iwai, H, Takeuchi, K, Kuzuhara, T, Komano, H, Kohda, D, Inagaki, F, Natori, S, Arata, Y, Shimada, I. | | Deposit date: | 2002-03-06 | | Release date: | 2002-03-27 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | 1H nuclear magnetic resonance study of the solution conformation of an antibacterial protein, sapecin.
FEBS Lett., 269, 1990
|
|
