4KDU
 
 | | Crystal structure of a glutathione transferase family member from Burkholderia graminis, target efi-507264, no gsh, ordered domains, space group P21, form(1) | | Descriptor: | Glutathione S-transferase domain | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-04-25 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a glutathione transferase family member from Burkholderia graminis, target efi-507264, no gsh, ordered domains, space group P21, form(1)
To be Published
|
|
4KEM
 
 | | Crystal structure of a tartrate dehydratase from azospirillum, target efi-502395, with bound mg and a putative acrylate ion, ordered active site | | Descriptor: | ACRYLIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Vetting, M.W, Wichelecki, D, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-04-25 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of a tartrate dehydratase from azospirillum, target efi-502395, with bound mg and a putative acrylate ion, ordered active site
To be Published
|
|
4NHZ
 
 | | Crystal structure of glutathione transferase BBTA-3750 from Bradyrhizobium sp., Target EFI-507290, with one glutathione bound | | Descriptor: | GLUTATHIONE, Putative glutathione S-transferase enzyme with thioredoxin-like domain | | Authors: | Patskovsky, Y, Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-11-05 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Crystal Structure of Glutathione Transferase Bbta-3750 from Bradyrhizobium Sp., Target Efi-507290
To be Published
|
|
4MF6
 
 | | Crystal structure of glutathione transferase BgramDRAFT_1843 from Burkholderia graminis, Target EFI-507289, with two glutathione molecules bound per one protein subunit | | Descriptor: | BENZOIC ACID, GLUTATHIONE, Glutathione S-transferase domain | | Authors: | Patskovsky, Y, Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-08-27 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of glutathione transferase BgramDRAFT_1843 from Burkholderia graminis, Target EFI-507289, with two glutathione molecules bound per one protein subunit
To be Published
|
|
4MF7
 
 | | Crystal structure of glutathione transferase BBTA-3750 from Bradyrhizobium sp., Target EFI-507290 | | Descriptor: | glutathione S-transferase enzyme with thioredoxin-like domain | | Authors: | Patskovsky, Y, Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-08-27 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of glutathione transferase BBTA-3750 from Bradyrhizobium sp., Target EFI-507290
To be Published
|
|
4NAV
 
 | | Crystal structure of hypothetical protein XCC2798 from Xanthomonas campestris, Target EFI-508608 | | Descriptor: | HYPOTHETICAL PROTEIN XCC279 | | Authors: | Kim, J, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Glenn, A.S, Chowdhury, S, Evans, B, Zhao, S.C, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Stead, M, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-22 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Crystal structure of hypothetical protein XCC2798 from Xanthomonas campestris, Target EFI-508608
TO BE PUBLISHED
|
|
3TS7
 
 | | CRYSTAL STRUCTURE OF FARNESYL DIPHOSPHATE SYNTHASE (TARGET EFI-501951) FROM Methylococcus capsulatus | | Descriptor: | Geranyltranstransferase, PHOSPHATE ION | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-09-12 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3UCA
 
 | | Crystal structure of isoprenoid synthase (target EFI-501974) from clostridium perfringens | | Descriptor: | Geranyltranstransferase | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-10-26 | | Release date: | 2011-11-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4DHD
 
 | | Crystal structure of isoprenoid synthase A3MSH1 (TARGET EFI-501992) from Pyrobaculum calidifontis | | Descriptor: | ACETATE ION, CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-01-27 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4GGB
 
 | | Crystal structure of a proposed galactarolactone cycloisomerase from agrobacterium tumefaciens, TARGET EFI-500704, WITH BOUND CA, DISORDERED LOOPS | | Descriptor: | CALCIUM ION, CHLORIDE ION, Putative uncharacterized protein | | Authors: | Vetting, M.W, Bouvier, J.T, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-08-06 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Purification, crystallization and structural elucidation of D-galactaro-1,4-lactone cycloisomerase from Agrobacterium tumefaciens involved in pectin degradation.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
4HPN
 
 | | Crystal structure of a proposed galactarolactone cycloisomerase from Agrobacterium Tumefaciens, target EFI-500704, with bound Ca, ordered loops | | Descriptor: | CALCIUM ION, IMIDAZOLE, NICKEL (II) ION, ... | | Authors: | Vetting, M.W, Bouvier, J.T, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-10-24 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Purification, crystallization and structural elucidation of D-galactaro-1,4-lactone cycloisomerase from Agrobacterium tumefaciens involved in pectin degradation.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
4H2H
 
 | | Crystal structure of an enolase (mandalate racemase subgroup, target EFI-502101) from Pelagibaca bermudensis htcc2601, with bound mg and l-4-hydroxyproline betaine (betonicine) | | Descriptor: | (2S,4R)-4-hydroxy-1,1-dimethylpyrrolidinium-2-carboxylate, (4S)-2-METHYL-2,4-PENTANEDIOL, IODIDE ION, ... | | Authors: | Vetting, M.W, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-09-12 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of new enzymes and metabolic pathways by using structure and genome context.
Nature, 502, 2013
|
|
4MHF
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Polaromonas sp. JS666 (Bpro_3107), target EFI-510173, with bound alpha/beta D-Glucuronate, space group P21 | | Descriptor: | TRAP dicarboxylate transporter, DctP subunit, alpha-D-glucopyranuronic acid, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Zhao, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-08-29 | | Release date: | 2013-09-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4MCO
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Rhodoferax ferrireducens (Rfer_1840), target EFI-510211, with bound malonate | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MALONATE ION, TRAP dicarboxylate transporter-DctP subunit | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Zhao, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-08-21 | | Release date: | 2013-09-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4MEV
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Rhodoferax ferrireducens (Rfer_1840), Target EFI-510211, with bound malonate, space group I422 | | Descriptor: | CITRIC ACID, MALONATE ION, TRAP dicarboxylate transporter-DctP subunit | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Zhao, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-08-27 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4LN5
 
 | | Crystal structure of a trap periplasmic solute binding protein from burkholderia ambifaria (Bamb_6123), TARGET EFI-510059, with bound glycerol and chloride ion | | Descriptor: | CHLORIDE ION, GLYCEROL, TRAP dicarboxylate transporter, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-07-11 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4MNC
 
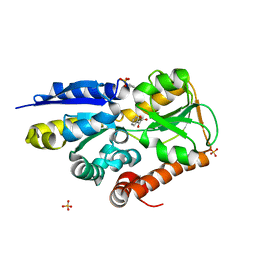 | | Crystal structure of a TRAP periplasmic solute binding protein from Polaromonas sp. JS666 (Bpro_4736), Target EFI-510156, with bound benzoyl formate, space group P21 | | Descriptor: | BENZOYL-FORMIC ACID, SULFATE ION, TRAP dicarboxylate transporter-DctP subunit | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Zhao, S, Stead, M, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-09-10 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4N8Y
 
 | | Crystal structure of a trap periplasmic solute binding protein from bradyrhizobium sp. btai1 b (bbta_0128), target EFI-510056 (bbta_0128), complex with alpha/beta-d-galacturonate | | Descriptor: | Putative TRAP-type C4-dicarboxylate transport system, binding periplasmic protein (DctP subunit), alpha-D-galactopyranuronic acid, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-18 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4N17
 
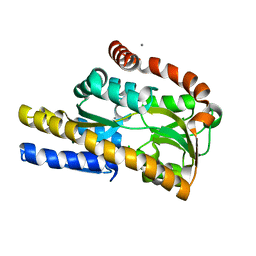 | | Crystal structure of a TRAP periplasmic solute binding protein from Burkholderia ambifaria (BAM_6123), Target EFI-510059, With bound beta-D-galacturonate | | Descriptor: | CALCIUM ION, CHLORIDE ION, TRAP dicarboxylate transporter, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Zhao, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-03 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4N6K
 
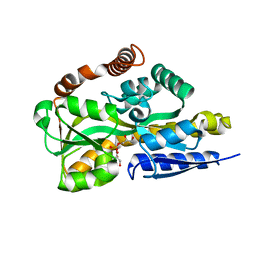 | | Crystal structure of a TRAP periplasmic solute binding protein from Desulfovibrio salexigens DSM2638, Target EFI-510113 (Desal_0342), complex with diglycerolphosphate | | Descriptor: | TRAP dicarboxylate transporter-DctP subunit, bis[(2S)-2,3-dihydroxypropyl] hydrogen phosphate | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-13 | | Release date: | 2013-11-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4N91
 
 | | Crystal structure of a trap periplasmic solute binding protein from anaerococcus prevotii dsm 20548 (Apre_1383), target EFI-510023, with bound alpha/beta d-glucuronate | | Descriptor: | CHLORIDE ION, TETRAETHYLENE GLYCOL, TRAP dicarboxylate transporter, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-18 | | Release date: | 2013-11-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4MX6
 
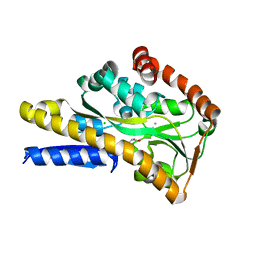 | | Crystal structure of a trap periplasmic solute binding protein from shewanella oneidensis (SO_3134), target EFI-510275, with bound succinate | | Descriptor: | CHLORIDE ION, SUCCINIC ACID, TRAP-type C4-dicarboxylate:H+ symport system substrate-binding component DctP | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Zhao, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-09-25 | | Release date: | 2013-10-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4N6D
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Desulfovibrio salexigens DSM2638 (Desal_3247), Target EFI-510112, phased with I3C, open complex, C-terminus of symmetry mate bound in ligand binding site | | Descriptor: | 1,2-ETHANEDIOL, 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, CHLORIDE ION, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-11 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4NGU
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Desulfovibrio alaskensis G20 (Dde_1548), Target EFI-510103, with bound D-Ala-D-Ala | | Descriptor: | CHLORIDE ION, D-ALANINE, SULFATE ION, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-11-02 | | Release date: | 2013-12-04 | | Last modified: | 2015-02-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4N4U
 
 | | Crystal structure of ABC transporter solute binding protein BB0719 from Bordetella bronchiseptica RB50, TARGET EFI-510049 | | Descriptor: | GLYCEROL, Putative ABC transporter periplasmic solute-binding protein | | Authors: | Patskovsky, Y, Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-08 | | Release date: | 2013-10-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
