3UBK
 
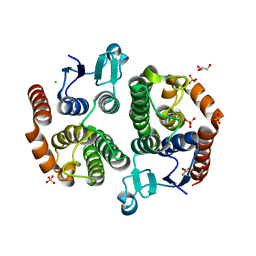 | | Crystal structure of glutathione transferase (TARGET EFI-501770) from leptospira interrogans | | Descriptor: | CHLORIDE ION, GLYCEROL, Glutathione transferase, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Zencheck, W.D, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-10-24 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Glutathione S-Transferase from Leptospira Interrogans
To be Published
|
|
3THU
 
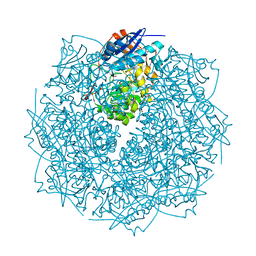 | | Crystal structure of an enolase from sphingomonas sp. ska58 (efi target efi-501683) with bound mg | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Wasserman, S.R, Morisco, L.L, Hillerich, B, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-08-19 | | Release date: | 2011-09-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of an enolase from sphingomonas sp. ska58 (efi target efi-501683) with bound mg
to be published
|
|
3TOU
 
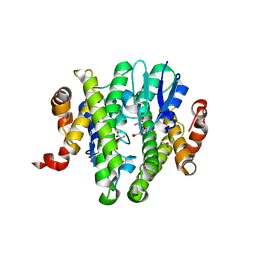 | | Crystal structure of GLUTATHIONE TRANSFERASE (TARGET EFI-501058) from Ralstonia solanacearum GMI1000 with GSH bound | | Descriptor: | ACETATE ION, GLUTATHIONE, Glutathione s-transferase protein | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Zencheck, W.D, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-09-06 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of GLUTATHIONE S-TRANSFERASE from Ralstonia solanacearum
To be Published
|
|
3UF0
 
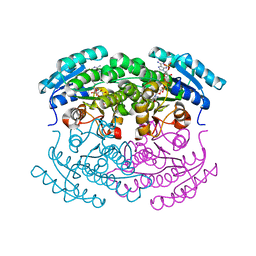 | | Crystal structure of a putative NAD(P) dependent gluconate 5-dehydrogenase from Beutenbergia cavernae(EFI target EFI-502044) with bound NADP (low occupancy) | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short-chain dehydrogenase/reductase SDR | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Hillerich, B, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-10-31 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a putative NAD(P) dependent gluconate 5-dehydrogenase from Beutenbergia cavernae(EFI target EFI-502044) with bound NADP (low occupancy)
To be Published
|
|
3UGV
 
 | | Crystal structure of an enolase from alpha pretobacterium bal199 (EFI TARGET EFI-501650) with bound MG | | Descriptor: | CHLORIDE ION, Enolase, MAGNESIUM ION, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Wasserman, S.R, Morisco, L.L, Hillerich, B, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-11-03 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of an enolase from alpha pretobacterium bal199 (EFI TARGET EFI-501650) with bound MG
to be published
|
|
3TTE
 
 | | Crystal structure of enolase brado_4202 (target EFI-501651) from Bradyrhizobium complexed with magnesium and mandelic acid | | Descriptor: | (S)-MANDELIC ACID, FORMIC ACID, GLYCEROL, ... | | Authors: | Patskovsky, Y, Kim, J, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammond, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-09-14 | | Release date: | 2011-09-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Mandelate Racemase from Bradyrhizobium Sp. Ors278
To be Published
|
|
3TOT
 
 | | Crystal structure of GLUTATHIONE TRANSFERASE (TARGET EFI-501058) from Ralstonia solanacearum GMI1000 | | Descriptor: | ACETATE ION, Glutathione s-transferase protein | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Zencheck, W.D, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-09-06 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of GLUTATHIONE S-TRANSFERASE from Ralstonia solanacearum
To be Published
|
|
3UJ2
 
 | | CRYSTAL STRUCTURE OF AN ENOLASE FROM ANAEROSTIPES CACCAE (EFI TARGET EFI-502054) WITH BOUND MG AND SULFATE | | Descriptor: | Enolase 1, MAGNESIUM ION, SULFATE ION | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Hillerich, B, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-11-07 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CRYSTAL STRUCTURE OF AN ENOLASE FROM ANAEROSTIPES CACCAE (EFI TARGET EFI-502054) WITH BOUND MG AND SULFATE
to be published
|
|
3V3W
 
 | | Crystal structure of an enolase from the soil bacterium Cellvibrio japonicus (TARGET EFI-502161) with bound MG and glycerol | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Wasserman, S.R, Morisco, L.L, Sojitra, S, Seidel, R, Hillerich, B, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-12-14 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of an enolase from the soil bacterium Cellvibrio japonicus (TARGET EFI-502161) with bound MG and glycerol
to be published
|
|
3V4B
 
 | | Crystal structure of an enolase from the soil bacterium Cellvibrio japonicus (TARGET EFI-502161) with bound MG and L-tartrate | | Descriptor: | CHLORIDE ION, L(+)-TARTARIC ACID, MAGNESIUM ION, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Wasserman, S.R, Morisco, L.L, Sojitra, S, Seidel, R.D, Hillerich, B, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Al Obaidi, N, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-12-14 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of an enolase from the soil bacterium Cellvibrio japonicus (TARGET EFI-502161) with bound MG and l-tartrate
to be published
|
|
4ECJ
 
 | | Crystal structure of glutathione s-transferase prk13972 (target efi-501853) from pseudomonas aeruginosa pacs2 complexed with glutathione | | Descriptor: | GLUTATHIONE, glutathione S-transferase | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Zencheck, W.D, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-03-26 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal Structure of Glutathione S-Transferase Prk13972 from Pseudomonas Aeruginosa
To be Published
|
|
4EXJ
 
 | | Crystal structure of glutathione s-transferase like protein lelg_03239 (target efi-501752) from lodderomyces elongisporus | | Descriptor: | CHLORIDE ION, SULFATE ION, uncharacterized protein | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Zencheck, W.D, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-04-30 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of glutathione s-transferase like protein lelg_03239 (target efi-501752) from lodderomyces elongisporus
To be Published
|
|
4EUM
 
 | | Crystal structure of a sugar kinase (Target EFI-502132) from Oceanicola granulosus with bound AMP, crystal form II | | Descriptor: | 2-dehydro-3-deoxygluconokinase, ADENOSINE MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-04-25 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a sugar kinase (Target EFI-502132) from Oceanicola granulosus with bound AMP, crystal form II
To be Published
|
|
4E69
 
 | | Crystal structure of a sugar kinase (target EFI-502132) from Oceanicola granulosus, unliganded structure | | Descriptor: | 1,2-ETHANEDIOL, 2-dehydro-3-deoxygluconokinase, CHLORIDE ION, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-03-15 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a sugar kinase (target EFI-502132) from Oceanicola granulosus, unliganded structure
To be Published
|
|
4EEK
 
 | | Crystal structure of HAD FAMILY HYDROLASE DR_1622 from Deinococcus radiodurans R1 (TARGET EFI-501256) With bound phosphate and sodium | | Descriptor: | Beta-phosphoglucomutase-related protein, PHOSPHATE ION, SODIUM ION | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Allen, K.N, Dunaway-Mariano, D, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-03-28 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of HAD HYDROLASE DR_1622 Deinococcus radiodurans R1 (TARGET EFI-501256)
To be Published
|
|
4DXK
 
 | | Crystal structure of an enolase (mandelate racemase subgroup, target EFI-502086) from Agrobacterium tumefaciens, with a succinimide residue, na and phosphate | | Descriptor: | Mandelate racemase / muconate lactonizing enzyme family protein, PHOSPHATE ION, SODIUM ION | | Authors: | Vetting, M.W, Bouvier, J.T, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-02-27 | | Release date: | 2012-04-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structure of an enolase (mandelate racemase subgroup, target EFI-502086) from Agrobacterium tumefaciens, with a succinimide residue, na and phosphate
to be published
|
|
4DZI
 
 | | Crystal structure of amidohydrolase map2389c (target EFI-500390) from Mycobacterium avium subsp. paratuberculosis K-10 | | Descriptor: | CADMIUM ION, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Raushel, F.M, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-03-01 | | Release date: | 2012-03-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of amidohydrolase map2389c (target EFI-500390) from Mycobacterium avium
To be Published
|
|
4DYE
 
 | | Crystal structure of an enolase (putative sugar isomerase, target efi-502095) from streptomyces coelicolor, no mg, ordered loop | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, isomerase | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-02-28 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of an enolase (putative sugar isomerase, target efi-502095) from streptomyces coelicolor, no mg, ordered loop
to be published
|
|
4DCC
 
 | | Crystal structure of had family enzyme bt-2542 from bacteroides thetaiotaomicron (target efi-501088) | | Descriptor: | CHLORIDE ION, Putative haloacid dehalogenase-like hydrolase, SODIUM ION, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Allen, K.N, Dunaway-Mariano, D, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-01-17 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of protein Bt-2542 from Bacteroides Thetaiotaomicron (Target Efi-501088)
To be Published
|
|
4DLM
 
 | | Crystal structure of an amidohydrolase (COG3618) from burkholderia multivorans (TARGET EFI-500235) with bound ZN, space group P212121 | | Descriptor: | Amidohydrolase 2, ZINC ION | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Seidel, R.D, Hillerich, B, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Al Obaidi, N.F, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Raushel, F.M, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-02-06 | | Release date: | 2012-02-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.925 Å) | | Cite: | Crystal structure of an amidohydrolase (COG3618) from burkholderia multivorans (TARGET EFI-500235) with bound ZN, space group P212121
to be published
|
|
4EZE
 
 | | Crystal structure of had family hydrolase t0658 from Salmonella enterica subsp. enterica serovar Typhi (Target EFI-501419) | | Descriptor: | CHLORIDE ION, Haloacid dehalogenase-like hydrolase, SODIUM ION | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Allen, K.N, Dunaway-Mariano, D, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-05-02 | | Release date: | 2012-07-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of had hydrolase t0658 from Salmonella enterica (Target EFI-501419)
To be Published
|
|
4F72
 
 | | Crystal structure of had family enzyme bt-2542 (target efi-501088) from Bacteroides thetaiotaomicron, asp12ala mutant, complex with magnesium and inorganic phosphate | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Patskovsky, Y, Farelli, J.D, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Zencheck, W.D, Gerlt, J.A, Allen, K.N, Dunaway-Mariano, D, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-05-15 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Protein Bt-2542 from Bacteroides Thetaiotaomicron (Target Efi-501088)
To be Published
|
|
4F71
 
 | | Crystal structure of had family enzyme bt-2542 (target efi-501088) from Bacteroides thetaiotaomicron, wild-type protein, complex with magnesium and inorganic phosphate | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Allen, K.N, Dunaway-Mariano, D, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-05-15 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal Structure of Protein Bt-2542 from Bacteroides Thetaiotaomicron (Target Efi-501088)
To be Published
|
|
4F4R
 
 | | Crystal structure of D-mannonate dehydratase homolog from Chromohalobacter salexigens (Target EFI-502114), with bound NA, ordered loop | | Descriptor: | CHLORIDE ION, D-mannonate dehydratase, GLYCEROL, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI), Medical Structural Genomics of Pathogenic Protozoa (MSGPP) | | Deposit date: | 2012-05-11 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of D-mannonate dehydratase homolog from Chromohalobacter salexigens (Target EFI-502114), with bound NA, ordered loop
to be published
|
|
4DX3
 
 | | Crystal structure of an enolase (mandelate racemase subgroup, target EFI-502086) from Agrobacterium tumefaciens, with a succinimide residue | | Descriptor: | CHLORIDE ION, GLYCEROL, Mandelate racemase / muconate lactonizing enzyme family protein | | Authors: | Vetting, M.W, Bouvier, J.T, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-02-27 | | Release date: | 2012-04-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of an enolase (mandelate racemase subgroup, target EFI-502086) from Agrobacterium tumefaciens, with a succinimide residue
to be published
|
|
