6IOU
 
 | | The ligand binding domain of Mlp24 with serine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Methyl-accepting chemotaxis protein, ... | | Authors: | Takahashi, Y, Sumita, K, Nishiyama, S, Kawagishi, I, Imada, K. | | Deposit date: | 2018-10-31 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Calcium Ions Modulate Amino Acid Sensing of the Chemoreceptor Mlp24 ofVibrio cholerae.
J. Bacteriol., 201, 2019
|
|
2ZVY
 
 | |
6IOR
 
 | | The ligand binding domain of Mlp24 with asparagine | | Descriptor: | ASPARAGINE, CALCIUM ION, Methyl-accepting chemotaxis protein | | Authors: | Takahashi, Y, Sumita, K, Nishiyama, S, Kawagishi, I, Imada, K. | | Deposit date: | 2018-10-31 | | Release date: | 2019-03-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Calcium Ions Modulate Amino Acid Sensing of the Chemoreceptor Mlp24 ofVibrio cholerae.
J. Bacteriol., 201, 2019
|
|
6IOT
 
 | | The ligand binding domain of Mlp24 with arginine | | Descriptor: | ARGININE, CALCIUM ION, Methyl-accepting chemotaxis protein | | Authors: | Takahashi, Y, Sumita, K, Nishiyama, S, Kawagishi, I, Imada, K. | | Deposit date: | 2018-10-31 | | Release date: | 2019-03-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Calcium Ions Modulate Amino Acid Sensing of the Chemoreceptor Mlp24 ofVibrio cholerae.
J. Bacteriol., 201, 2019
|
|
6IOS
 
 | | The ligand binding domain of Mlp24 with proline | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Takahashi, Y, Sumita, K, Nishiyama, S, Kawagishi, I, Imada, K. | | Deposit date: | 2018-10-31 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Calcium Ions Modulate Amino Acid Sensing of the Chemoreceptor Mlp24 ofVibrio cholerae.
J. Bacteriol., 201, 2019
|
|
7CMG
 
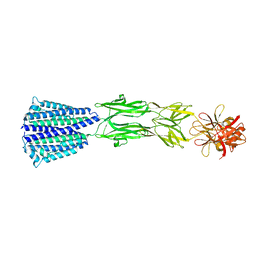 | | The Structure of the periplasmic domain of PorM | | Descriptor: | Por secretion system protein porM/gldM | | Authors: | Sato, K, Okada, K, Nakayam, K, Imada, K. | | Deposit date: | 2020-07-27 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | PorM, a core component of bacterial type IX secretion system, forms a dimer with a unique kinked-rod shape.
Biochem.Biophys.Res.Commun., 532, 2020
|
|
6IOP
 
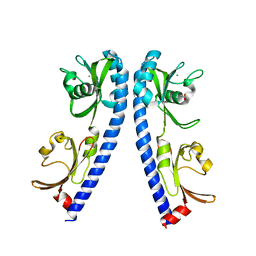 | | The ligand binding domain of Mlp24 | | Descriptor: | ACETATE ION, ALANINE, CALCIUM ION, ... | | Authors: | Sumita, K, Takahashi, Y, Nishiyama, S, Kawagishi, I, Imada, K. | | Deposit date: | 2018-10-31 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Calcium Ions Modulate Amino Acid Sensing of the Chemoreceptor Mlp24 ofVibrio cholerae.
J. Bacteriol., 201, 2019
|
|
6JF2
 
 | |
3ATP
 
 | | Structure of the ligand binding domain of the bacterial serine chemoreceptor Tsr with ligand | | Descriptor: | Methyl-accepting chemotaxis protein I, SERINE | | Authors: | Tajima, H, Sakuma, M, Homma, K, Kawagishi, I, Imada, K. | | Deposit date: | 2011-01-07 | | Release date: | 2011-10-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ligand specificity determined by differentially arranged common ligand-binding residues in bacterial amino acid chemoreceptors Tsr and Tar.
J.Biol.Chem., 286, 2011
|
|
6IOV
 
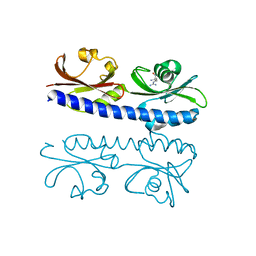 | | The ligand binding domain of Mlp37 with arginine | | Descriptor: | ARGININE, Methyl-accepting chemotaxis (MCP) signaling domain protein | | Authors: | Takahashi, Y, Sumita, K, Nishiyama, S, Kawagishi, I, Imada, K. | | Deposit date: | 2018-10-31 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.351 Å) | | Cite: | Structural basis of the binding affinity of chemoreceptors Mlp24p and Mlp37p for various amino acids.
Biochem.Biophys.Res.Commun., 523, 2020
|
|
7D84
 
 | | 34-fold symmetry Salmonella S ring formed by full-length FliF | | Descriptor: | Flagellar M-ring protein | | Authors: | Kawamoto, A, Miyata, T, Makino, F, Kinoshita, M, Minamino, T, Imada, K, Kato, T, Namba, K. | | Deposit date: | 2020-10-07 | | Release date: | 2021-05-19 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Native flagellar MS ring is formed by 34 subunits with 23-fold and 11-fold subsymmetries.
Nat Commun, 12, 2021
|
|
3X0V
 
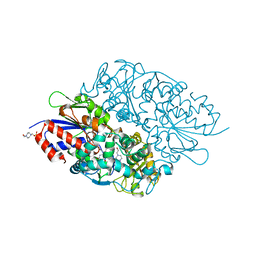 | | Structure of L-lysine oxidase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, L-lysine oxidase | | Authors: | Sano, T, Uchida, Y, Amano, M, Kawaguchi, T, Kondo, H, Inagaki, K, Imada, K. | | Deposit date: | 2014-10-22 | | Release date: | 2015-04-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Recombinant expression, molecular characterization and crystal structure of antitumor enzyme, l-lysine alpha-oxidase from Trichoderma viride.
J.Biochem., 157, 2015
|
|
7C3I
 
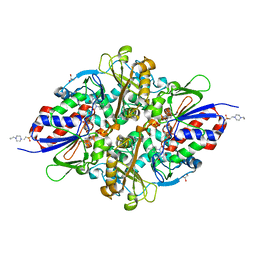 | | Structure of L-lysine oxidase D212A/D315A | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Kitagawa, M, Matsumoto, Y, Inagaki, K, Imada, K. | | Deposit date: | 2020-05-12 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of strict substrate recognition of l-lysine alpha-oxidase from Trichoderma viride.
Protein Sci., 29, 2020
|
|
7C3L
 
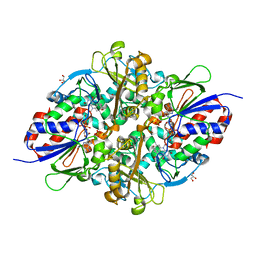 | | Structure of L-lysine oxidase D212A/D315A in complex with L-tyrosine | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, L-lysine oxidase, ... | | Authors: | Kitagawa, M, Matsumoto, Y, Inagaki, K, Imada, K. | | Deposit date: | 2020-05-12 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of strict substrate recognition of l-lysine alpha-oxidase from Trichoderma viride.
Protein Sci., 29, 2020
|
|
7DMA
 
 | |
7DM9
 
 | |
7EHA
 
 | | Crystal structure of the flagellar hook cap from Salmonella enterica serovar Typhimurium | | Descriptor: | Basal-body rod modification protein FlgD | | Authors: | Matsunami, H, Yoon, Y.-H, Imada, K, Namba, K, Samatey, F.A. | | Deposit date: | 2021-03-29 | | Release date: | 2021-11-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the bacterial flagellar hook cap provides insights into a hook assembly mechanism
Commun Biol, 4, 2021
|
|
7EH9
 
 | | Crystal structure of the flagellar hook cap fragment from Salmonella enterica serovar Typhimurium | | Descriptor: | Basal-body rod modification protein FlgD | | Authors: | Matsunami, H, Yoon, Y.-H, Imada, K, Namba, K, Samatey, F.A. | | Deposit date: | 2021-03-29 | | Release date: | 2021-11-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the bacterial flagellar hook cap provides insights into a hook assembly mechanism
Commun Biol, 4, 2021
|
|
7E0D
 
 | | Structure of L-glutamate oxidase R305E mutant in complex with L-arginine | | Descriptor: | ARGININE, FLAVIN-ADENINE DINUCLEOTIDE, L-glutamate oxidase | | Authors: | Ito, N, Matsuo, S, Inagaki, K, Imada, K. | | Deposit date: | 2021-01-27 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A new l-arginine oxidase engineered from l-glutamate oxidase.
Protein Sci., 30, 2021
|
|
7E0C
 
 | | Structure of L-glutamate oxidase R305E mutant | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, L-glutamate oxidase | | Authors: | Ito, N, Matsuo, S, Inagaki, K, Imada, K. | | Deposit date: | 2021-01-27 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A new l-arginine oxidase engineered from l-glutamate oxidase.
Protein Sci., 30, 2021
|
|
7D4D
 
 | | Structure of L-lysine oxidase precursor in complex with L-lysine (1.24M) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, L-Lysine alpha-oxidase, LYSINE | | Authors: | Kitagawa, M, Ito, N, Matsumoto, Y, Inagaki, K, Imada, K. | | Deposit date: | 2020-09-23 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural basis of enzyme activity regulation by the propeptide of l-lysine alpha-oxidase precursor from Trichoderma viride .
J Struct Biol X, 5, 2021
|
|
7D4E
 
 | | Structure of L-lysine oxidase precursor in complex with L-lysine (1.0 M) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, L-Lysine alpha-oxidase, LYSINE | | Authors: | Kitagawa, M, Ito, N, Matsumoto, Y, Inagaki, K, Imada, K. | | Deposit date: | 2020-09-23 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis of enzyme activity regulation by the propeptide of l-lysine alpha-oxidase precursor from Trichoderma viride .
J Struct Biol X, 5, 2021
|
|
7D4C
 
 | | Structure of L-lysine oxidase precursor | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, L-Lysine alpha-oxidase, PHOSPHATE ION | | Authors: | Ito, N, Kitagawa, M, Matsumoto, Y, Inagaki, K, Imada, K. | | Deposit date: | 2020-09-23 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural basis of enzyme activity regulation by the propeptide of l-lysine alpha-oxidase precursor from Trichoderma viride .
J Struct Biol X, 5, 2021
|
|
7CIK
 
 | | Structure of the 58-213 fragment of FliF | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Flagellar M-ring protein | | Authors: | Takekawa, T, Sakuma, M, Kojima, S, Homma, M, Imada, K. | | Deposit date: | 2020-07-07 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Two Distinct Conformations in 34 FliF Subunits Generate Three Different Symmetries within the Flagellar MS-Ring.
Mbio, 12, 2021
|
|
7C3H
 
 | | Structure of L-lysine oxidase in complex with L-lysine | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Kondo, H, Kitagawa, M, Sugiyama, S, Imada, K. | | Deposit date: | 2020-05-12 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of strict substrate recognition of l-lysine alpha-oxidase from Trichoderma viride.
Protein Sci., 29, 2020
|
|
