1BE5
 
 | | STRUCTURAL STUDIES OF A STABLE PARALLEL-STRANDED DNA DUPLEX INCORPORATING ISOGUANINE:CYTOSINE AND ISOCYTOSINE:GUANINE BASE PAIRS BY NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DNA DUPLEX (TGCACGGACT) | | Authors: | Yang, X.-L, Sugiyama, H, Ikeda, S, Saito, I, Wang, A.H.-J. | | Deposit date: | 1998-05-19 | | Release date: | 1998-08-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural studies of a stable parallel-stranded DNA duplex incorporating isoguanine:cytosine and isocytosine:guanine basepairs by nuclear magnetic resonance spectroscopy.
Biophys.J., 75, 1998
|
|
2N9K
 
 | | 1H, 13C, and 15N Chemical Shift Assignments for in vitro GB1 | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Ikeya, T, Hanashima, T, Hosoya, S, Shimazaki, M, Ikeda, S, Mishima, M, Guentert, P, Ito, Y. | | Deposit date: | 2015-11-26 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Improved in-cell structure determination of proteins at near-physiological concentration
Sci Rep, 6, 2016
|
|
2N9L
 
 | | 1H, 13C, and 15N Chemical Shift Assignments for in-cell GB1 | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Ikeya, T, Hanashima, T, Hosoya, S, Shimazaki, M, Ikeda, S, Mishima, M, Guentert, P, Ito, Y. | | Deposit date: | 2015-11-30 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Improved in-cell structure determination of proteins at near-physiological concentration
Sci Rep, 6, 2016
|
|
3CV9
 
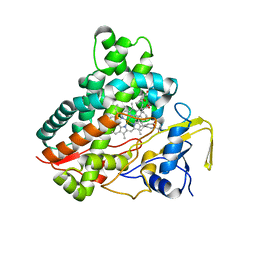 | | Crystal structure of vitamin D hydroxylase cytochrome P450 105A1 (R73A/R84A mutant) in complex with 1alpha,25-dihydroxyvitamin D3 | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, Cytochrome P450-SU1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hayashi, K, Sugimoto, H, Shinkyo, R, Yamada, M, Ikeda, S, Ikushiro, S, Kamakura, M, Shiro, Y, Sakaki, T. | | Deposit date: | 2008-04-18 | | Release date: | 2008-11-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-based design of a highly active vitamin D hydroxylase from Streptomyces griseolus CYP105A1
Biochemistry, 47, 2008
|
|
3CV8
 
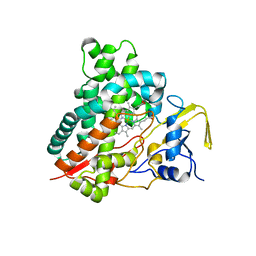 | | Crystal structure of vitamin D hydroxylase cytochrome P450 105A1 (R84F mutant) | | Descriptor: | Cytochrome P450-SU1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hayashi, K, Sugimoto, H, Shinkyo, R, Yamada, M, Ikeda, S, Ikushiro, S, Kamakura, M, Shiro, Y, Sakaki, T. | | Deposit date: | 2008-04-18 | | Release date: | 2008-11-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design of a highly active vitamin D hydroxylase from Streptomyces griseolus CYP105A1
Biochemistry, 47, 2008
|
|
