7S6S
 
 | | Complex structure of Methane monooxygenase hydroxylase and regulatory subunit DBL1 | | 分子名称: | 1,2-ETHANEDIOL, BENZOIC ACID, FE (III) ION, ... | | 著者 | Johns, J.C, Banerjee, R, Semonis, M.M, Shi, K, Aihara, H, Lipscomb, J.D. | | 登録日 | 2021-09-14 | | 公開日 | 2021-12-29 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | X-ray Crystal Structures of Methane Monooxygenase Hydroxylase Complexes with Variants of Its Regulatory Component: Correlations with Altered Reaction Cycle Dynamics.
Biochemistry, 61, 2022
|
|
8EUO
 
 | | Hydroxynitrile Lyase from Hevea brasiliensis with Seven Mutations | | 分子名称: | (S)-hydroxynitrile lyase | | 著者 | Greenberg, L.R, Walsh, M.E, Kazlauskas, R.J, Pierce, C.T, Shi, K, Aihara, H, Evans, R.L. | | 登録日 | 2022-10-19 | | 公開日 | 2022-11-30 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | to be published
To Be Published
|
|
6VOY
 
 | | Cryo-EM structure of HTLV-1 instasome | | 分子名称: | DNA (25-MER), DNA (5'-D(P*AP*CP*AP*CP*AP*CP*TP*TP*GP*AP*CP*TP*AP*GP*GP*GP*TP*G)-3'), DNA-binding protein 7d, ... | | 著者 | Bhatt, V, Shi, K, Sundborger, A, Aihara, H. | | 登録日 | 2020-02-01 | | 公開日 | 2020-07-01 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural basis of host protein hijacking in human T-cell leukemia virus integration.
Nat Commun, 11, 2020
|
|
1C0N
 
 | | CSDB PROTEIN, NIFS HOMOLOGUE | | 分子名称: | ACETIC ACID, PROTEIN (CSDB PROTEIN), PYRIDOXAL-5'-PHOSPHATE | | 著者 | Fujii, T, Maeda, M, Mihara, H, Kurihara, T, Esaki, N, Hata, Y. | | 登録日 | 1999-07-17 | | 公開日 | 2000-07-17 | | 最終更新日 | 2017-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure of a NifS homologue: X-ray structure analysis of CsdB, an Escherichia coli counterpart of mammalian selenocysteine lyase
Biochemistry, 39, 2000
|
|
6JR0
 
 | | Crystal structure of the human nucleosome phased with 12 selenium atoms | | 分子名称: | CHLORIDE ION, DNA (146-MER), Histone H2A type 1-B/E, ... | | 著者 | Saotome, M, Horikoshi, N, Urano, K, Kujirai, T, Yuzurihara, H, Kurumizaka, H, Kagawa, W. | | 登録日 | 2019-04-02 | | 公開日 | 2019-10-02 | | 最終更新日 | 2019-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure determination of the nucleosome core particle by selenium SAD phasing.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
8E14
 
 | | Cryo-EM structure of Rous sarcoma virus strand transfer complex | | 分子名称: | DNA (42-MER), DNA (5'-D(*AP*AP*TP*GP*TP*TP*GP*TP*CP*TP*TP*AP*TP*GP*CP*AP*AP*TP*AP*CP*TP*C)-3'), DNA (5'-D(*AP*GP*TP*GP*TP*CP*TP*TP*CP*TP*TP*CP*TP*TP*TP*C)-3'), ... | | 著者 | Pandey, K.K, Bera, S, Shi, K, Aihara, H, Grandgenett, D.P. | | 登録日 | 2022-08-09 | | 公開日 | 2023-04-26 | | 最終更新日 | 2023-10-18 | | 実験手法 | ELECTRON MICROSCOPY (3.36 Å) | | 主引用文献 | Molecular determinants for Rous sarcoma virus intasome assemblies involved in retroviral integration.
J.Biol.Chem., 299, 2023
|
|
7K30
 
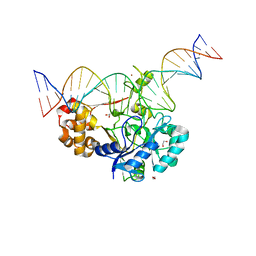 | | Crystal structure of Endonuclease Q complex with 27-mer duplex substrate with dU at the active site | | 分子名称: | 1,2-ETHANEDIOL, DNA (27-MER), Endonuclease Q, ... | | 著者 | Shi, K, Moeller, N.M, Banerjee, S, Yin, L, Orellana, K, Aihara, H. | | 登録日 | 2020-09-10 | | 公開日 | 2021-03-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.34 Å) | | 主引用文献 | Structural basis for recognition of distinct deaminated DNA lesions by endonuclease Q.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7K31
 
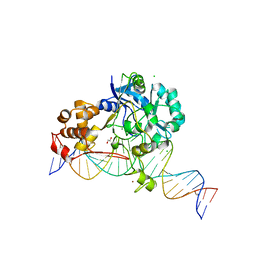 | | Crystal structure of Endonuclease Q complex with 27-mer duplex substrate with dI at the active site | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA (27-MER), ... | | 著者 | Shi, K, Moeller, N.M, Banerjee, S, Yin, L, Orellana, K, Aihara, H. | | 登録日 | 2020-09-10 | | 公開日 | 2021-03-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.88 Å) | | 主引用文献 | Structural basis for recognition of distinct deaminated DNA lesions by endonuclease Q.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7RWN
 
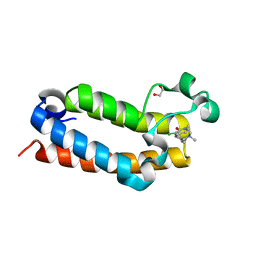 | | Crystal Structure of BPTF bromodomain in complex with 4-chloro-5-{4-[(dimethylamino)methyl]anilino}-2-methylpyridazin-3(2H)-one | | 分子名称: | 1,2-ETHANEDIOL, 4-chloro-5-{4-[(dimethylamino)methyl]anilino}-2-methylpyridazin-3(2H)-one, Nucleosome-remodeling factor subunit BPTF | | 著者 | Zahid, H, Buchholz, C, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | 登録日 | 2021-08-20 | | 公開日 | 2022-08-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.39 Å) | | 主引用文献 | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
6JR1
 
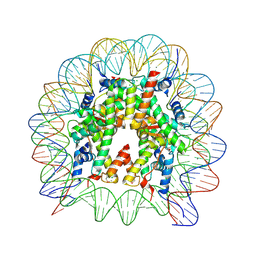 | | Crystal structure of the human nucleosome phased with 16 selenium atoms | | 分子名称: | CHLORIDE ION, DNA (146-MER), Histone H2A type 1-B/E, ... | | 著者 | Saotome, M, Horikoshi, N, Urano, K, Kujirai, T, Yuzurihara, H, Kurumizaka, H, Kagawa, W. | | 登録日 | 2019-04-02 | | 公開日 | 2019-10-02 | | 最終更新日 | 2019-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structure determination of the nucleosome core particle by selenium SAD phasing.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
7RWP
 
 | | Crystal Structure of BPTF bromodomain in complex with 5-[4-(aminomethyl)anilino]-4-chloro-2-methylpyridazin-3(2H)-one | | 分子名称: | 5-[4-(aminomethyl)anilino]-4-chloro-2-methylpyridazin-3(2H)-one, CALCIUM ION, Nucleosome-remodeling factor subunit BPTF | | 著者 | Zahid, H, Buchholz, C, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | 登録日 | 2021-08-20 | | 公開日 | 2022-08-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
7RWQ
 
 | | Crystal Structure of BPTF bromodomain in complex with 4-chloro-2-methyl-5-[(1,2,3,4-tetrahydroisoquinolin-6-yl)amino]pyridazin-3(2H)-one | | 分子名称: | 4-chloro-2-methyl-5-[(1,2,3,4-tetrahydroisoquinolin-6-yl)amino]pyridazin-3(2H)-one, CALCIUM ION, Nucleosome-remodeling factor subunit BPTF | | 著者 | Zahid, H, Buchholz, C, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | 登録日 | 2021-08-20 | | 公開日 | 2022-08-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
7RWO
 
 | | Crystal Structure of BPTF bromodomain in complex with 4-chloro-2-methyl-5-[(1,2,3,4-tetrahydroisoquinolin-7-yl)amino]pyridazin-3(2H)-one | | 分子名称: | 1,2-ETHANEDIOL, 4-chloro-2-methyl-5-[(1,2,3,4-tetrahydroisoquinolin-7-yl)amino]pyridazin-3(2H)-one, Nucleosome-remodeling factor subunit BPTF | | 著者 | Zahid, H, Buchholz, C, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | 登録日 | 2021-08-20 | | 公開日 | 2022-08-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
6VK7
 
 | | Crystal Structure of reduced Methylosinus trichosporium OB3b Soluble Methane Monooxygenase Hydroxylase | | 分子名称: | FE (III) ION, Methane monooxygenase, Methane monooxygenase component A alpha chain | | 著者 | Jones, J.C, Banerjee, R, Shi, K, Aihara, H, Lipscomb, J.D. | | 登録日 | 2020-01-18 | | 公開日 | 2020-08-05 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | Structural Studies of theMethylosinus trichosporiumOB3b Soluble Methane Monooxygenase Hydroxylase and Regulatory Component Complex Reveal a Transient Substrate Tunnel.
Biochemistry, 59, 2020
|
|
5U0L
 
 | | X-ray crystal structure of fatty aldehyde dehydrogenase enzymes from Marinobacter aquaeolei VT8 complexed with a substrate | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | 著者 | Shi, K, Mulliner, K, Barney, B.M, Aihara, H. | | 登録日 | 2016-11-24 | | 公開日 | 2017-04-26 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.285 Å) | | 主引用文献 | Five Fatty Aldehyde Dehydrogenase Enzymes from Marinobacter and Acinetobacter spp. and Structural Insights into the Aldehyde Binding Pocket.
Appl. Environ. Microbiol., 83, 2017
|
|
6VK6
 
 | | Crystal Structure of Methylosinus trichosporium OB3b Soluble Methane Monooxygenase Hydroxylase | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, FE (III) ION, ... | | 著者 | Jones, J.C, Banerjee, R, Shi, K, Aihara, H, Lipscomb, J.D. | | 登録日 | 2020-01-18 | | 公開日 | 2020-08-05 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | Structural Studies of theMethylosinus trichosporiumOB3b Soluble Methane Monooxygenase Hydroxylase and Regulatory Component Complex Reveal a Transient Substrate Tunnel.
Biochemistry, 59, 2020
|
|
5U0M
 
 | | Fatty aldehyde dehydrogenase from Marinobacter aquaeolei VT8 and cofactor complex | | 分子名称: | 1,2-ETHANEDIOL, CITRIC ACID, N-succinylglutamate 5-semialdehyde dehydrogenase, ... | | 著者 | Shi, K, Mulliner, K, Barney, B.M, Aihara, H. | | 登録日 | 2016-11-24 | | 公開日 | 2017-04-26 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.075 Å) | | 主引用文献 | Five Fatty Aldehyde Dehydrogenase Enzymes from Marinobacter and Acinetobacter spp. and Structural Insights into the Aldehyde Binding Pocket.
Appl. Environ. Microbiol., 83, 2017
|
|
1Z1G
 
 | | Crystal structure of a lambda integrase tetramer bound to a Holliday junction | | 分子名称: | 25-MER, 29-MER, 5'-D(*AP*CP*AP*GP*GP*TP*CP*AP*CP*TP*AP*TP*CP*AP*GP*TP*CP*AP*AP*AP*AP*TP*AP*CP*C)-3', ... | | 著者 | Biswas, T, Aihara, H, Radman-Livaja, M, Filman, D, Landy, A, Ellenberger, T. | | 登録日 | 2005-03-03 | | 公開日 | 2005-06-28 | | 最終更新日 | 2021-10-20 | | 実験手法 | X-RAY DIFFRACTION (4.4 Å) | | 主引用文献 | A structural basis for allosteric control of DNA recombination by lambda integrase.
Nature, 435, 2005
|
|
6CWJ
 
 | | Crystal structures of cyanuric acid hydrolase from Moorella thermoacetica complexed with 1,3-Acetone Dicarboxylic Acid | | 分子名称: | 1,3-PROPANDIOL, 3-oxopentanedioic acid, ACETATE ION, ... | | 著者 | Shi, K, Aihara, H. | | 登録日 | 2018-03-30 | | 公開日 | 2019-06-12 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.253 Å) | | 主引用文献 | Crystal structures of Moorella thermoacetica cyanuric acid hydrolase reveal conformational flexibility and asymmetry important for catalysis.
Plos One, 14, 2019
|
|
8E5D
 
 | | Crystal structure of double-stranded DNA deaminase toxin DddA in complex with DNA with the target cytosine parked in the major groove | | 分子名称: | DNA (5'-D(*GP*TP*AP*CP*CP*GP*GP*AP*CP*GP*TP*TP*GP*C)-3'), Double-stranded DNA deaminase toxin A, MAGNESIUM ION, ... | | 著者 | Yin, L.L, Shi, K, Aihara, H. | | 登録日 | 2022-08-21 | | 公開日 | 2023-05-24 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | Structural basis of sequence-specific cytosine deamination by double-stranded DNA deaminase toxin DddA.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8SPH
 
 | | Crystal structure of chimeric omicron RBD (strain XBB.1) complexed with human ACE2 | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Zhang, W, Shi, K, Aihara, H, Li, F. | | 登録日 | 2023-05-03 | | 公開日 | 2023-08-02 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.71 Å) | | 主引用文献 | Structural evolution of SARS-CoV-2 omicron in human receptor recognition.
J.Virol., 97, 2023
|
|
6DRT
 
 | |
8EJO
 
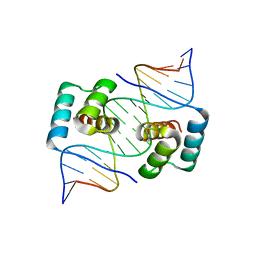 | | Crystal structure of the homeodomain of Platypus sDUX in complex with DNA | | 分子名称: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*CP*GP*TP*AP*AP*TP*CP*TP*AP*AP*TP*CP*AP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*TP*GP*AP*TP*TP*AP*GP*AP*TP*TP*AP*CP*GP*C)-3'), ... | | 著者 | Yin, L.L, Shi, K, Aihara, H. | | 登録日 | 2022-09-18 | | 公開日 | 2023-09-20 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.67 Å) | | 主引用文献 | Antagonism among DUX family members evolved from an ancestral toxic single homeodomain protein.
Iscience, 26, 2023
|
|
8EJP
 
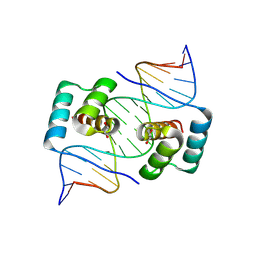 | | Crystal structure of the homeodomain of Platypus sDUX in complex with DNA containing 5-Bromouracil | | 分子名称: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*CP*GP*TP*AP*AP*TP*CP*TP*AP*AP*TP*CP*AP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*TP*GP*AP*TP*TP*AP*GP*AP*TP*TP*AP*CP*GP*C)-3'), ... | | 著者 | Yin, L.L, Shi, K, Aihara, H. | | 登録日 | 2022-09-18 | | 公開日 | 2023-09-20 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.174 Å) | | 主引用文献 | Antagonism among DUX family members evolved from an ancestral toxic single homeodomain protein.
Iscience, 26, 2023
|
|
8SBM
 
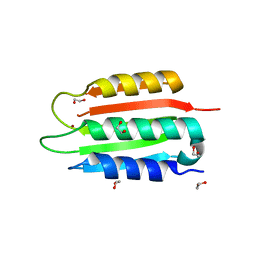 | | Crystal structure of the wild-type Catalytic ATP-binding domain of Mtb DosS | | 分子名称: | 1,2-ETHANEDIOL, GAF domain-containing protein, SODIUM ION, ... | | 著者 | Larson, G, Shi, K, Aihara, H, Bhagi-Damodaran, A. | | 登録日 | 2023-04-03 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 | Understanding ATP Binding to DosS Catalytic Domain with a Short ATP-Lid.
Biochemistry, 62, 2023
|
|
