4GN9
 
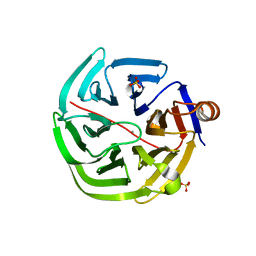 | | mouse SMP30/GNL-glucose complex | | Descriptor: | CALCIUM ION, Regucalcin, SULFATE ION, ... | | Authors: | Aizawa, S, Senda, M, Harada, A, Maruyama, N, Ishida, T, Aigaki, T, Ishigami, A, Senda, T. | | Deposit date: | 2012-08-17 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of the gamma-lactone-ring formation in ascorbic acid biosynthesis by the senescence marker protein-30/gluconolactonase
Plos One, 8, 2013
|
|
1Q3S
 
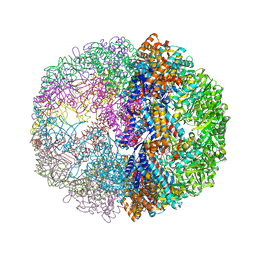 | | Crystal structure of the chaperonin from Thermococcus strain KS-1 (FormIII crystal complexed with ADP) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Thermosome alpha subunit | | Authors: | Shomura, Y, Yoshida, T, Iizuka, R, Maruyama, T, Yohda, M, Miki, K. | | Deposit date: | 2003-07-31 | | Release date: | 2004-01-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structures of the Group II Chaperonin from Thermococcus strain KS-1: Steric Hindrance by the Substituted Amino Acid, and Inter-subunit Rearrangement between Two Crystal Forms.
J.Mol.Biol., 335, 2004
|
|
1Q3R
 
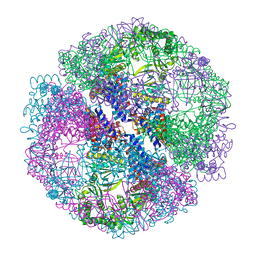 | | Crystal structure of the chaperonin from Thermococcus strain KS-1 (nucleotide-free form of single mutant) | | Descriptor: | SULFATE ION, Thermosome alpha subunit | | Authors: | Shomura, Y, Yoshida, T, Iizuka, R, Maruyama, T, Yohda, M, Miki, K. | | Deposit date: | 2003-07-31 | | Release date: | 2004-01-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structures of the Group II Chaperonin from Thermococcus strain KS-1: Steric Hindrance by the Substituted Amino Acid, and Inter-subunit Rearrangement between Two Crystal Forms.
J.Mol.Biol., 335, 2004
|
|
1Q3Q
 
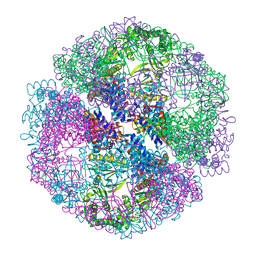 | | Crystal structure of the chaperonin from Thermococcus strain KS-1 (two-point mutant complexed with AMP-PNP) | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Thermosome alpha subunit | | Authors: | Shomura, Y, Yoshida, T, Iizuka, R, Maruyama, T, Yohda, M, Miki, K. | | Deposit date: | 2003-07-31 | | Release date: | 2004-01-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of the Group II Chaperonin from Thermococcus strain KS-1: Steric Hindrance by the Substituted Amino Acid, and Inter-subunit Rearrangement between Two Crystal Forms.
J.Mol.Biol., 335, 2004
|
|
1Q2V
 
 | | Crystal structure of the chaperonin from Thermococcus strain KS-1 (nucleotide-free form) | | Descriptor: | SULFATE ION, Thermosome alpha subunit | | Authors: | Shomura, Y, Yoshida, T, Iizuka, R, Yohda, M, Maruyama, T, Miki, K. | | Deposit date: | 2003-07-26 | | Release date: | 2004-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of the Group II Chaperonin from Thermococcus strain KS-1: Steric Hindrance by the Substituted Amino Acid, and Inter-subunit Rearrangement between Two Crystal Forms.
J.Mol.Biol., 335, 2004
|
|
3VJL
 
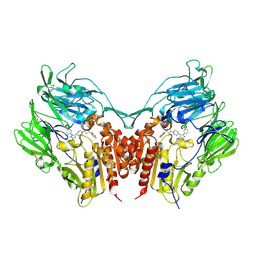 | | Crystal structure of human depiptidyl peptidase IV (DPP-4) in complex with a prolylthiazolidine inhibitor #2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, ... | | Authors: | Akahoshi, F, Kishida, H, Miyaguchi, I, Yoshida, T, Ishii, S. | | Deposit date: | 2011-10-24 | | Release date: | 2012-10-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Discovery and preclinical profile of teneligliptin (3-[(2S,4S)-4-[4-(3-methyl-1-phenyl-1H-pyrazol-5-yl)piperazin-1-yl]pyrrolidin-2-ylcarbonyl]thiazolidine): A highly potent, selective, long-lasting and orally active dipeptidyl peptidase IV inhibitor for the treatment of type 2 diabetes
Bioorg.Med.Chem., 20, 2012
|
|
3VJM
 
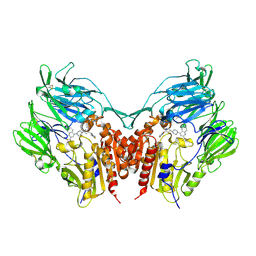 | | Crystal structure of human depiptidyl peptidase IV (DPP-4) in complex with a prolylthiazolidine inhibitor #1 | | Descriptor: | 1,3-thiazolidin-3-yl[(2S,4S)-4-{4-[2-(trifluoromethyl)quinolin-4-yl]piperazin-1-yl}pyrrolidin-2-yl]methanone, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Akahoshi, F, Kishida, H, Miyaguchi, I, Yoshida, T, Ishii, S. | | Deposit date: | 2011-10-24 | | Release date: | 2012-08-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Fused bicyclic heteroarylpiperazine-substituted l-prolylthiazolidines as highly potent DPP-4 inhibitors lacking the electrophilic nitrile group
Bioorg.Med.Chem., 20, 2012
|
|
2ELG
 
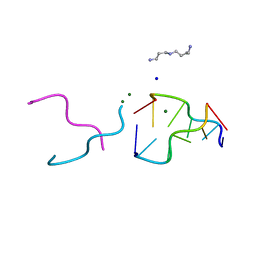 | | The rare crystallographic structure of d(CGCGCG)2: The natural spermidine molecule bound to the minor groove of left-handed Z-DNA d(CGCGCG)2 at 10 degree celsius | | Descriptor: | DNA (5'-D(*DCP*DGP*DCP*DGP*DCP*DG)-3'), MAGNESIUM ION, SODIUM ION, ... | | Authors: | Ohishi, H, Tozuka, Y, Zhou, D.Y, Ishida, T, Nakatani, K. | | Deposit date: | 2007-03-27 | | Release date: | 2008-04-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | The rare crystallographic structure of d(CGCGCG)(2): The natural spermidine molecule bound to the minor groove of left-handed Z-DNA d(CGCGCG)(2) at 10 degrees C
Biochem.Biophys.Res.Commun., 358, 2007
|
|
1RTU
 
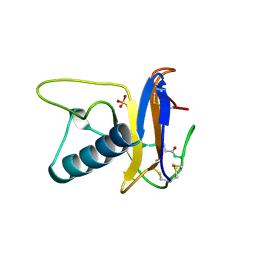 | | USTILAGO SPHAEROGENA RIBONUCLEASE U2 | | Descriptor: | RIBONUCLEASE U2, SULFATE ION | | Authors: | Noguchi, S, Satow, Y, Uchida, T, Sasaki, C, Matsuzaki, T. | | Deposit date: | 1995-05-12 | | Release date: | 1996-11-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Ustilago sphaerogena ribonuclease U2 at 1.8 A resolution.
Biochemistry, 34, 1995
|
|
1PE6
 
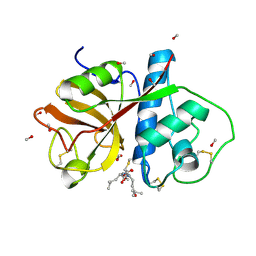 | | REFINED X-RAY STRUCTURE OF PAPAIN(DOT)E-64-C COMPLEX AT 2.1-ANGSTROMS RESOLUTION | | Descriptor: | METHANOL, N-[1-HYDROXYCARBOXYETHYL-CARBONYL]LEUCYLAMINO-2-METHYL-BUTANE, PAPAIN | | Authors: | Yamamoto, D, Matsumoto, K, Ohishi, H, Ishida, T, Inoue, M, Kitamura, K, Mizuno, H. | | Deposit date: | 1991-05-14 | | Release date: | 1993-04-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Refined x-ray structure of papain.E-64-c complex at 2.1-A resolution.
J.Biol.Chem., 266, 1991
|
|
2Z48
 
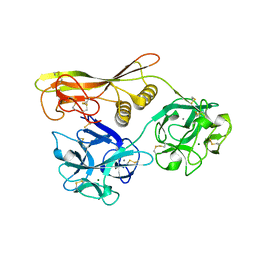 | | Crystal Structure of Hemolytic Lectin CEL-III Complexed with GalNac | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-galactopyranose, CALCIUM ION, ... | | Authors: | Hatakeyama, T, Unno, H, Eto, S, Hidemura, H, Uchida, T, Kouzuma, Y. | | Deposit date: | 2007-06-13 | | Release date: | 2007-10-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | C-type lectin-like carbohydrate-recognition of the hemolytic lectin CEL-III containing ricin-type beta-trefoil folds
J.Biol.Chem., 282, 2007
|
|
3ANU
 
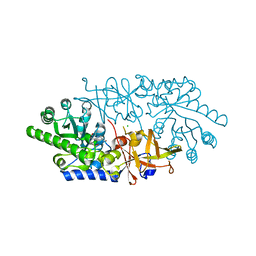 | | Crystal structure of D-serine dehydratase from chicken kidney | | Descriptor: | CHLORIDE ION, D-serine dehydratase, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Tanaka, H, Senda, M, Venugopalan, N, Yamamoto, A, Senda, T, Ishida, T, Horiike, K. | | Deposit date: | 2010-09-09 | | Release date: | 2011-06-15 | | Last modified: | 2013-08-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a zinc-dependent D-serine dehydratase from chicken kidney
J.Biol.Chem., 286, 2011
|
|
3B2C
 
 | | Crystal structure of the collagen triple helix model [{PRO-HYP(R)-GLY}4-{HYP(S)-Pro-GLY}2-{PRO-HYP(R)-GLY}4]3 | | Descriptor: | Collagen-like peptide | | Authors: | Motooka, D, Kawahara, K, Nakamura, S, Doi, M, Nishi, Y, Nishiuchi, Y, Nakazawa, T, Yoshida, T, Ohkubo, T, Kobayashi, Y, Kang, Y.K, Uchiyama, S. | | Deposit date: | 2011-07-26 | | Release date: | 2012-04-04 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | The triple helical structure and stability of collagen model peptide with 4(S)-hydroxyprolyl-pro-gly units
Biopolymers, 98, 2011
|
|
2ZUF
 
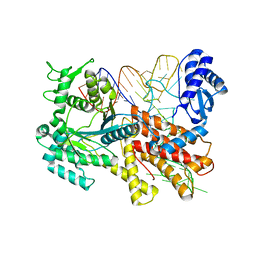 | | Crystal structure of Pyrococcus horikoshii arginyl-tRNA synthetase complexed with tRNA(Arg) | | Descriptor: | Arginyl-tRNA synthetase, tRNA-Arg | | Authors: | Konno, M, Sumida, T, Uchikawa, E, Mori, Y, Yanagisawa, T, Sekine, S, Yokoyama, S. | | Deposit date: | 2008-10-16 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Modeling of tRNA-assisted mechanism of Arg activation based on a structure of Arg-tRNA synthetase, tRNA, and an ATP analog (ANP)
Febs J., 276, 2009
|
|
2ZUE
 
 | | Crystal structure of Pyrococcus horikoshii arginyl-tRNA synthetase complexed with tRNA(Arg) and an ATP analog (ANP) | | Descriptor: | Arginyl-tRNA synthetase, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Konno, M, Sumida, T, Uchikawa, E, Mori, Y, Yanagisawa, T, Sekine, S, Yokoyama, S. | | Deposit date: | 2008-10-16 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Modeling of tRNA-assisted mechanism of Arg activation based on a structure of Arg-tRNA synthetase, tRNA, and an ATP analog (ANP)
Febs J., 276, 2009
|
|
2Z49
 
 | | Crystal Structure of Hemolytic Lectin CEL-III Complexed with methyl-alpha-D-galactopylanoside | | Descriptor: | CALCIUM ION, Hemolytic lectin CEL-III, MAGNESIUM ION, ... | | Authors: | Hatakeyama, T, Unno, H, Eto, S, Hidemura, H, Uchida, T, Kouzuma, Y. | | Deposit date: | 2007-06-13 | | Release date: | 2007-10-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | C-type lectin-like carbohydrate-recognition of the hemolytic lectin CEL-III containing ricin-type beta-trefoil folds
J.Biol.Chem., 282, 2007
|
|
3AFV
 
 | | Dye-decolorizing peroxidase (DyP) at 1.4 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DyP, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugano, Y, Yoshida, T, Tsuge, H. | | Deposit date: | 2010-03-11 | | Release date: | 2011-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The catalytic mechanism of dye-decolorizing peroxidase DyP may require the swinging movement of an aspartic acid residue
Febs J., 278, 2011
|
|
3ANV
 
 | | Crystal structure of D-serine dehydratase from chicken kidney (2,3-DAP complex) | | Descriptor: | 3-amino-D-alanine, CHLORIDE ION, D-serine dehydratase, ... | | Authors: | Tanaka, H, Senda, M, Venugopalan, N, Yamamoto, A, Senda, T, Ishida, T, Horiike, K. | | Deposit date: | 2010-09-09 | | Release date: | 2011-06-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of a zinc-dependent D-serine dehydratase from chicken kidney
J.Biol.Chem., 286, 2011
|
|
1PIP
 
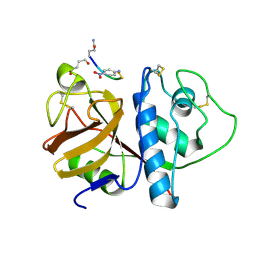 | | CRYSTAL STRUCTURE OF PAPAIN-SUCCINYL-GLN-VAL-VAL-ALA-ALA-P-NITROANILIDE COMPLEX AT 1.7 ANGSTROMS RESOLUTION: NONCOVALENT BINDING MODE OF A COMMON SEQUENCE OF ENDOGENOUS THIOL PROTEASE INHIBITORS | | Descriptor: | Papain, SUCCINYL-GLN-VAL-VAL-ALA-ALA-P-NITROANILIDE | | Authors: | Yamamoto, A, Tomoo, K, Doi, M, Ohishi, H, Inoue, M, Ishida, T, Yamamoto, D, Tsuboi, S, Okamoto, H, Okada, Y. | | Deposit date: | 1992-10-03 | | Release date: | 1993-10-31 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of papain-succinyl-Gln-Val-Val-Ala-Ala-p-nitroanilide complex at 1.7-A resolution: noncovalent binding mode of a common sequence of endogenous thiol protease inhibitors.
Biochemistry, 31, 1992
|
|
3AWN
 
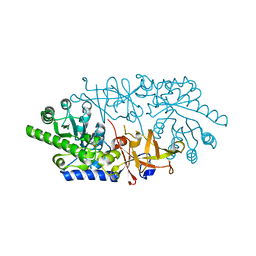 | | Crystal structure of D-serine dehydratase from chicken kidney (EDTA treated) | | Descriptor: | D-serine dehydratase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Tanaka, H, Senda, M, Venugopalan, N, Yamamoto, A, Senda, T, Ishida, T, Horiike, K. | | Deposit date: | 2011-03-25 | | Release date: | 2011-06-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a zinc-dependent D-serine dehydratase from chicken kidney.
J.Biol.Chem., 286, 2011
|
|
3AWO
 
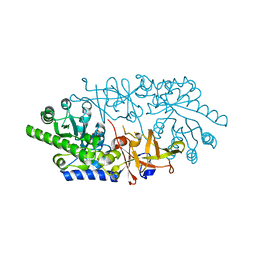 | | Crystal structure of D-serine dehydratase in complex with D-serine from chicken kidney (EDTA-treated) | | Descriptor: | D-SERINE, D-serine dehydratase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Tanaka, H, Senda, M, Venugopalan, N, Yamamoto, A, Senda, T, Ishida, T, Horiike, K. | | Deposit date: | 2011-03-25 | | Release date: | 2011-06-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of a zinc-dependent D-serine dehydratase from chicken kidney.
J.Biol.Chem., 286, 2011
|
|
3VOR
 
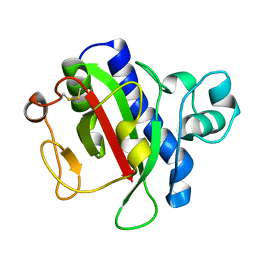 | | Crystal Structure Analysis of the CofA | | Descriptor: | CFA/III pilin | | Authors: | Fukakusa, S, Kawahara, K, Nakamura, S, Iwasita, T, Baba, S, Nishimura, M, Kobayashi, Y, Honda, T, Iida, T, Taniguchi, T, Ohkubo, T. | | Deposit date: | 2012-02-06 | | Release date: | 2012-09-26 | | Last modified: | 2013-07-31 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Structure of the CFA/III major pilin subunit CofA from human enterotoxigenic Escherichia coli determined at 0.90 A resolution by sulfur-SAD phasing
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2ZZJ
 
 | | Crystal structure of endo-beta-1,4-glucuronan lyase from fungus Trichoderma reesei | | Descriptor: | CALCIUM ION, CITRIC ACID, Glucuronan lyase A | | Authors: | Konno, N, Ishida, T, Fushinobu, S, Igarashi, K, Habu, N, Samejima, M, Isogai, A. | | Deposit date: | 2009-02-16 | | Release date: | 2009-05-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of polysaccharide lyase family 20 endo-beta-1,4-glucuronan lyase from the filamentous fungus Trichoderma reesei.
Febs Lett., 583, 2009
|
|
3B1L
 
 | |
3X07
 
 | | Crystal structure of PIP4KIIBETA N203A complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Takeuchi, K, Lo, Y.H, Sumita, K, Senda, M, Terakawa, J, Dimitoris, A, Locasale, J.W, Sasaki, M, Yoshino, H, Zhang, Y, Kahoud, E.R, Takano, T, Yokota, T, Emerling, B, Asara, J.A, Ishida, T, Shimada, I, Daikoku, T, Cantley, L.C, Senda, T, Sasaki, A.T. | | Deposit date: | 2014-10-09 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Lipid Kinase PI5P4K beta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis
Mol.Cell, 61, 2016
|
|
