3WHR
 
 | | Crystal structure of gamma-glutamyltranspeptidase from Bacillus subtilis (crystal soaked for 3min. in acivicin soln. ) | | Descriptor: | Gamma-glutamyltranspeptidase large chain, Gamma-glutamyltranspeptidase small chain | | Authors: | Ida, T, Suzuki, H, Fukuyama, K, Hiratake, J, Wada, K. | | Deposit date: | 2013-08-30 | | Release date: | 2014-02-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structure of Bacillus subtilis gamma-glutamyltranspeptidase in complex with acivicin: diversity of the binding mode of a classical and electrophilic active-site-directed glutamate analogue.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8K2K
 
 | | Crystal structure of Group 3 Oligosaccharide/Monosaccharide-releasing beta-N-acetylgalactosaminidase NgaDssm in complex with GalNAc-thiazoline | | Descriptor: | (3aR,5R,6R,7R,7aR)-5-(hydroxymethyl)-2-methyl-5,6,7,7a-tetrahydro-3aH-pyrano[3,2-d][1,3]thiazole-6,7-diol, CHLORIDE ION, GLYCEROL, ... | | Authors: | Sumida, T, Fushinobu, S. | | Deposit date: | 2023-07-12 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Genetic and functional diversity of beta-N-acetylgalactosamine-targeting glycosidases expanded by deep-sea metagenome analysis.
Nat Commun, 15, 2024
|
|
8K2J
 
 | |
8K2L
 
 | |
8K2G
 
 | |
8K2I
 
 | | Crystal structure of Group 2 Oligosaccharide/Monosaccharide-releasing beta-N-acetylhexosaminidase NgaAt from Arabidopsis thaliana in complex with GlcNAc-thiazoline | | Descriptor: | 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sumida, T, Fushinobu, S. | | Deposit date: | 2023-07-12 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Genetic and functional diversity of beta-N-acetylgalactosamine-targeting glycosidases expanded by deep-sea metagenome analysis.
Nat Commun, 15, 2024
|
|
8K2H
 
 | | Crystal structure of Group 2Oligosaccharide/Monosaccharide-releasing beta-N-acetylhexosaminidase NgaAt from Arabidopsis thaliana in complex with GalNAc-thiazoline | | Descriptor: | (3aR,5R,6R,7R,7aR)-5-(hydroxymethyl)-2-methyl-5,6,7,7a-tetrahydro-3aH-pyrano[3,2-d][1,3]thiazole-6,7-diol, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sumida, T, Fushinobu, S. | | Deposit date: | 2023-07-12 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Genetic and functional diversity of beta-N-acetylgalactosamine-targeting glycosidases expanded by deep-sea metagenome analysis.
Nat Commun, 15, 2024
|
|
8K2M
 
 | | Crystal structure of Group 4 Monosaccharide-releasing beta-N-acetylgalactosaminidase NgaP2 from Paenibacillus sp. TS12 in complex with GalNAc-thiazoline | | Descriptor: | (3aR,5R,6R,7R,7aR)-5-(hydroxymethyl)-2-methyl-5,6,7,7a-tetrahydro-3aH-pyrano[3,2-d][1,3]thiazole-6,7-diol, BROMIDE ION, Monosaccharide-releasing beta-N-acetylgalactosaminidase | | Authors: | Sumida, T, Fushinobu, S. | | Deposit date: | 2023-07-12 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Genetic and functional diversity of beta-N-acetylgalactosamine-targeting glycosidases expanded by deep-sea metagenome analysis.
Nat Commun, 15, 2024
|
|
8K2N
 
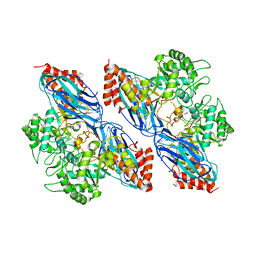 | |
8K2F
 
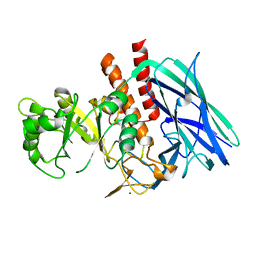 | |
7OLG
 
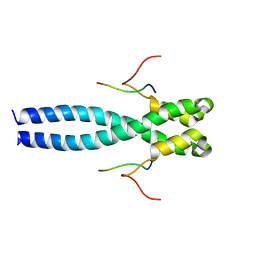 | | EB1 bound to MACF peptide | | Descriptor: | 11MACF, Microtubule-associated protein RP/EB family member 1 | | Authors: | Almeida, T.B, Barsukov, I.L. | | Deposit date: | 2021-05-20 | | Release date: | 2022-06-01 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | EB1 bound to MACF peptide
To Be Published
|
|
1GE9
 
 | | SOLUTION STRUCTURE OF THE RIBOSOME RECYCLING FACTOR | | Descriptor: | RIBOSOME RECYCLING FACTOR | | Authors: | Yoshida, T, Uchiyama, S, Nakano, H, Kashimori, H, Kijima, H, Ohshima, T, Saihara, Y, Ishino, T, Shimahara, T, Yoshida, T, Yokose, K, Ohkubo, T, Kaji, A, Kobayashi, Y. | | Deposit date: | 2000-10-19 | | Release date: | 2001-05-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the ribosome recycling factor from Aquifex aeolicus.
Biochemistry, 40, 2001
|
|
7VTJ
 
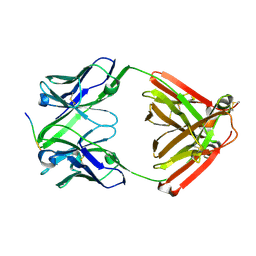 | | The cross-reaction complex structure with VQIIYK peptide and tau antibody's Fab domain. | | Descriptor: | Heavy chain of Fab, Light chain of Fab, VQIIYK peptide | | Authors: | Tsuchida, T, Fukuhara, N, Tsuchiya, T, Miyamoto, K, In, Y, Minoura, K, Taniguchi, Y, Ishida, T, Tomoo, K. | | Deposit date: | 2021-10-29 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The cross-reaction complex structure with VQIIYK peptide and tau antibody's Fab domain.
To Be Published
|
|
6KXX
 
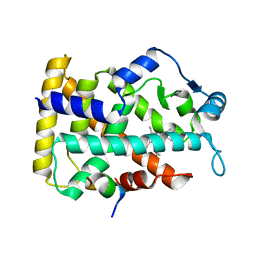 | | Human PPAR alpha ligand binding domain in complex with a synthetic agonist (compound A) | | Descriptor: | 1-(4-chlorophenyl)-6-methyl-3-propan-2-yl-pyrazolo[3,4-b]pyridine-4-carboxylic acid, PGC1alpha, Peroxisome proliferator-activated receptor alpha | | Authors: | Yoshida, T, Tachibana, K, Oki, H, Doi, M, Fukuda, S, Yuzuriha, T, Tabata, R, Ishimoto, K, Kawahara, K, Ohkubo, T, Miyachi, H, Doi, T. | | Deposit date: | 2019-09-14 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Basis for PPAR alpha Activation by 1H-pyrazolo-[3,4-b]pyridine Derivatives.
Sci Rep, 10, 2020
|
|
6KXY
 
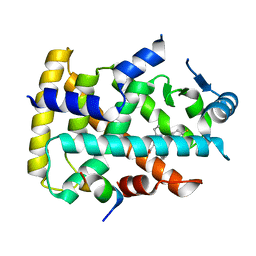 | | Human PPAR alpha ligand binding domain in complex with a synthetic agonist (compound B) | | Descriptor: | 6-ethyl-1-(4-fluorophenyl)-3-pentan-3-yl-pyrazolo[3,4-b]pyridine-4-carboxylic acid, PGC1alpha, Peroxisome proliferator-activated receptor alpha | | Authors: | Yoshida, T, Tachibana, K, Oki, H, Doi, M, Fukuda, S, Yuzuriha, T, Tabata, R, Ishimoto, K, Kawahara, K, Ohkubo, T, Miyachi, H, Doi, T. | | Deposit date: | 2019-09-14 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for PPAR alpha Activation by 1H-pyrazolo-[3,4-b]pyridine Derivatives.
Sci Rep, 10, 2020
|
|
6KLW
 
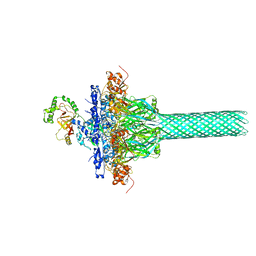 | | Complex structure of Iota toxin enzymatic component (Ia) and binding component (Ib) pore with long stem | | Descriptor: | CALCIUM ION, Iota toxin component Ia, Iota toxin component Ib | | Authors: | Yoshida, T, Yamada, T, Kawamoto, A, Mitsuoka, K, Iwasaki, K, Tsuge, H. | | Deposit date: | 2019-07-30 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures reveal translocational unfolding in the clostridial binary iota toxin complex.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6KLX
 
 | | Pore structure of Iota toxin binding component (Ib) | | Descriptor: | CALCIUM ION, Iota toxin component Ib | | Authors: | Yoshida, T, Yamada, T, Kawamoto, A, Mitsuoka, K, Iwasaki, K, Tsuge, H. | | Deposit date: | 2019-07-30 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures reveal translocational unfolding in the clostridial binary iota toxin complex.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6KLO
 
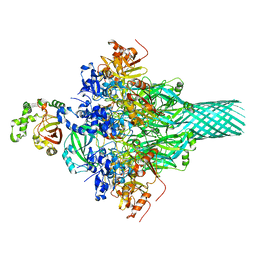 | | Complex structure of Iota toxin enzymatic component (Ia) and binding component (Ib) pore with short stem | | Descriptor: | CALCIUM ION, Iota toxin component Ia, Iota toxin component Ib | | Authors: | Yoshida, T, Yamada, T, Kawamoto, A, Mitsuoka, K, Iwasaki, K, Tsuge, H. | | Deposit date: | 2019-07-30 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures reveal translocational unfolding in the clostridial binary iota toxin complex.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5C2I
 
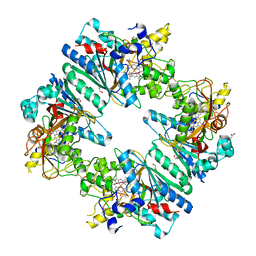 | | Crystal structure of Anabaena sp. DyP-type peroxidese (AnaPX) | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Alr1585 protein, ... | | Authors: | Yoshida, T, Amano, Y, Tsuge, H, Sugano, Y. | | Deposit date: | 2015-06-16 | | Release date: | 2015-12-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Anabaena sp. DyP-type peroxidase is a tetramer consisting of two asymmetric dimers.
Proteins, 84, 2016
|
|
7FGK
 
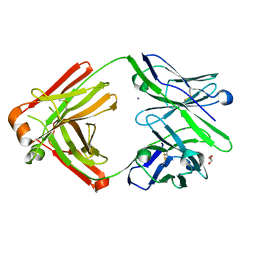 | | The Fab antibody single structure against tau protein. | | Descriptor: | Fab Heavy Chain, Fab Light Chain, GLYCEROL, ... | | Authors: | Tsuchida, T, Susa, K, Kibiki, T, Tsuchiya, T, Miyamoto, K, In, Y, Minoura, K, Taniguchi, T, Ishida, T, Tomoo, K. | | Deposit date: | 2021-07-27 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The free structure of the Fab domain of antibody that recognizes the PHF core region VQIINK in Tau protein.
To Be Published
|
|
7FGL
 
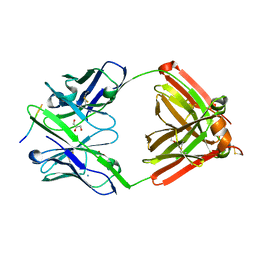 | | The complex structure of PHF core domain peptide of tau, VQIVYK, and antibody's Fab domain. | | Descriptor: | AMMONIUM ION, Fab Heavy Chain, Fab Light Chain, ... | | Authors: | Tsuchida, T, Tsuchiya, T, Miyamoto, K, In, Y, Minoura, K, Taniguchi, T, Ishida, T, Tomoo, K. | | Deposit date: | 2021-07-27 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The cross-reaction complex structure with VQIVYK of tau and the antibody's Fab domain.
To Be Published
|
|
6JX3
 
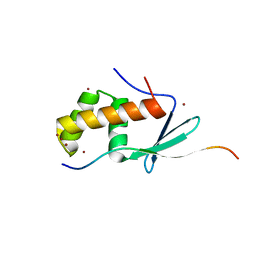 | |
7FGJ
 
 | | The cross-reaction complex structure with VQILNK peptide and the tau antibody's Fab domain. | | Descriptor: | CHLORIDE ION, Fab Heavy Chain, Fab Light Chain, ... | | Authors: | Tsuchida, T, Tsuchiya, T, Miyamoto, K, In, Y, Minoura, K, Taniguchi, T, Ishida, T, Tomoo, K. | | Deposit date: | 2021-07-27 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The cross-reaction complex structure with VQILNK peptide and the antibody's Fab domain.
To Be Published
|
|
7FGR
 
 | | The cross-reaction complex structure with VQIFNK peptide and the tau antibody's Fab domain. | | Descriptor: | AMMONIUM ION, CHLORIDE ION, Fab Heavy Chain, ... | | Authors: | Tsuchida, T, Tsuchiya, T, Miyamoto, K, In, Y, Minoura, K, Taniguchi, T, Ishida, T, Tomoo, K. | | Deposit date: | 2021-07-27 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The cross-reaction complex structure with VQIFNK peptide and the tau antibody's Fab domain.
To Be Published
|
|
1PPP
 
 | |
