3SUV
 
 | |
3SUR
 
 | | Crystal structure of beta-hexosaminidase from Paenibacillus sp. TS12 in complex with NAG-thiazoline. | | 分子名称: | 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, Beta-hexosaminidase, SULFATE ION | | 著者 | Sumida, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2011-07-11 | | 公開日 | 2012-06-06 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Gaining insight into the inhibition of glycoside hydrolase family 20 exo-beta-N-acetylhexosaminidases using a structural approach
Org.Biomol.Chem., 10, 2012
|
|
3SUU
 
 | | Crystal structure of beta-hexosaminidase from Paenibacillus sp. TS12 in complex with Gal-PUGNAc | | 分子名称: | Beta-hexosaminidase, SULFATE ION, [(Z)-[(3R,4R,5R,6R)-3-acetamido-6-(hydroxymethyl)-4,5-bis(oxidanyl)oxan-2-ylidene]amino] N-phenylcarbamate | | 著者 | Sumida, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2011-07-11 | | 公開日 | 2012-06-06 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Gaining insight into the inhibition of glycoside hydrolase family 20 exo-beta-N-acetylhexosaminidases using a structural approach
Org.Biomol.Chem., 10, 2012
|
|
3SUT
 
 | |
5C2I
 
 | | Crystal structure of Anabaena sp. DyP-type peroxidese (AnaPX) | | 分子名称: | 1,2-ETHANEDIOL, ACETIC ACID, Alr1585 protein, ... | | 著者 | Yoshida, T, Amano, Y, Tsuge, H, Sugano, Y. | | 登録日 | 2015-06-16 | | 公開日 | 2015-12-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Anabaena sp. DyP-type peroxidase is a tetramer consisting of two asymmetric dimers.
Proteins, 84, 2016
|
|
5DE0
 
 | | Dye-decolorizing protein from V. cholerae | | 分子名称: | Deferrochelatase, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Uchida, T, Sasaki, M, Tanaka, Y, Yao, M. | | 登録日 | 2015-08-25 | | 公開日 | 2015-10-21 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | A Dye-Decolorizing Peroxidase from Vibrio cholerae.
Biochemistry, 54, 2015
|
|
3EQN
 
 | | Crystal structure of beta-1,3-glucanase from Phanerochaete chrysosporium (Lam55A) | | 分子名称: | ACETATE ION, GLYCEROL, Glucan 1,3-beta-glucosidase, ... | | 著者 | Ishida, T, Fushinobu, S, Kawai, R, Kitaoka, M, Igarashi, K, Samejima, M. | | 登録日 | 2008-10-01 | | 公開日 | 2009-02-03 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of glycoside hydrolase family 55 beta -1,3-glucanase from the basidiomycete Phanerochaete chrysosporium
J.Biol.Chem., 284, 2009
|
|
3GH5
 
 | | Crystal structure of beta-hexosaminidase from Paenibacillus sp. TS12 in complex with GlcNAc | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, beta-hexosaminidase | | 著者 | Sumida, T, Ishii, R, Yanagisawa, T, Yokoyama, S, Ito, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2009-03-03 | | 公開日 | 2009-07-07 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Molecular cloning and crystal structural analysis of a novel beta-N-acetylhexosaminidase from Paenibacillus sp. TS12 capable of degrading glycosphingolipids
J.Mol.Biol., 392, 2009
|
|
3GH7
 
 | | Crystal structure of beta-hexosaminidase from Paenibacillus sp. TS12 in complex with GalNAc | | 分子名称: | 2-acetamido-2-deoxy-beta-D-galactopyranose, SULFATE ION, beta-hexosaminidase | | 著者 | Sumida, T, Ishii, R, Yanagisawa, T, Yokoyama, S, Ito, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2009-03-03 | | 公開日 | 2009-07-07 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Molecular cloning and crystal structural analysis of a novel beta-N-acetylhexosaminidase from Paenibacillus sp. TS12 capable of degrading glycosphingolipids
J.Mol.Biol., 392, 2009
|
|
3GH4
 
 | | Crystal structure of beta-hexosaminidase from Paenibacillus sp. TS12 | | 分子名称: | ACETIC ACID, SULFATE ION, beta-hexosaminidase | | 著者 | Sumida, T, Ishii, R, Yanagisawa, T, Yokoyama, S, Ito, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2009-03-03 | | 公開日 | 2009-07-07 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Molecular cloning and crystal structural analysis of a novel beta-N-acetylhexosaminidase from Paenibacillus sp. TS12 capable of degrading glycosphingolipids
J.Mol.Biol., 392, 2009
|
|
1VCL
 
 | | Crystal Structure of Hemolytic Lectin CEL-III | | 分子名称: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Uchida, T, Yamasaki, T, Eto, S, Sugawara, H, Kurisu, G, Nakagawa, A, Kusunoki, M, Hatakeyama, T. | | 登録日 | 2004-03-09 | | 公開日 | 2004-09-07 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal Structure of the Hemolytic Lectin CEL-III Isolated from the Marine Invertebrate Cucumaria echinata: IMPLICATIONS OF DOMAIN STRUCTURE FOR ITS MEMBRANE PORE-FORMATION MECHANISM
J.Biol.Chem., 279, 2004
|
|
3EQO
 
 | | Crystal structure of beta-1,3-glucanase from Phanerochaete chrysosporium (Lam55A) gluconolactone complex | | 分子名称: | D-glucono-1,5-lactone, Glucan 1,3-beta-glucosidase, ZINC ION, ... | | 著者 | Ishida, T, Fushinobu, S, Kawai, R, Kitaoka, M, Igarashi, K, Samejima, M. | | 登録日 | 2008-10-01 | | 公開日 | 2009-02-03 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Crystal structure of glycoside hydrolase family 55 beta -1,3-glucanase from the basidiomycete Phanerochaete chrysosporium
J.Biol.Chem., 284, 2009
|
|
5ZJ4
 
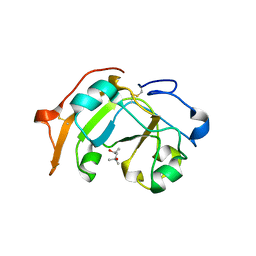 | | Guanine-specific ADP-ribosyltransferase | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADP-ribosyltransferase | | 著者 | Yoshida, T, Tsuge, H. | | 登録日 | 2018-03-19 | | 公開日 | 2018-08-08 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.49739277 Å) | | 主引用文献 | Substrate N2atom recognition mechanism in pierisin family DNA-targeting, guanine-specific ADP-ribosyltransferase ScARP.
J. Biol. Chem., 293, 2018
|
|
5ZJ5
 
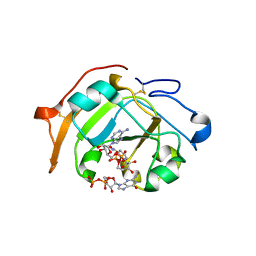 | | Guanine-specific ADP-ribosyltransferase with NADH and GDP | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ADP-ribosyltransferase, GUANOSINE-5'-DIPHOSPHATE | | 著者 | Yoshida, T, Tsuge, H. | | 登録日 | 2018-03-19 | | 公開日 | 2018-08-08 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.568112 Å) | | 主引用文献 | Substrate N2atom recognition mechanism in pierisin family DNA-targeting, guanine-specific ADP-ribosyltransferase ScARP.
J. Biol. Chem., 293, 2018
|
|
1PPP
 
 | |
3A5Z
 
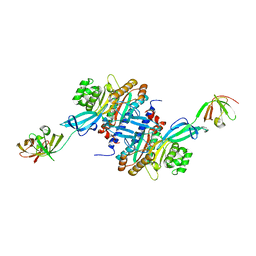 | | Crystal structure of Escherichia coli GenX in complex with elongation factor P | | 分子名称: | 5'-O-[(L-LYSYLAMINO)SULFONYL]ADENOSINE, Elongation factor P, Putative lysyl-tRNA synthetase | | 著者 | Sumida, T, Yanagisawa, T, Ishii, R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2009-08-17 | | 公開日 | 2010-08-25 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | A paralog of lysyl-tRNA synthetase aminoacylates a conserved lysine residue in translation elongation factor P.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3A5Y
 
 | | Crystal structure of GenX from Escherichia coli in complex with lysyladenylate analog | | 分子名称: | 5'-O-[(L-LYSYLAMINO)SULFONYL]ADENOSINE, Putative lysyl-tRNA synthetase | | 著者 | Sumida, T, Yanagisawa, T, Ishii, R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2009-08-17 | | 公開日 | 2010-08-25 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | A paralog of lysyl-tRNA synthetase aminoacylates a conserved lysine residue in translation elongation factor P.
Nat.Struct.Mol.Biol., 17, 2010
|
|
6JWF
 
 | | Holo form of Pyranose Dehydrogenase PQQ domain from Coprinopsis cinerea | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | 著者 | Takeda, K, Ishida, T, Yoshida, M, Samejima, M, Ohno, H, Igarashi, K, Nakamura, N. | | 登録日 | 2019-04-20 | | 公開日 | 2019-11-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Crystal Structure of the Catalytic and CytochromebDomains in a Eukaryotic Pyrroloquinoline Quinone-Dependent Dehydrogenase.
Appl.Environ.Microbiol., 85, 2019
|
|
6JT5
 
 | | Crystal structure of PQQ doamin of Pyranose Dehydrogenase from Coprinopsis cinerea: apo-from | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Extracellular PQQ-dependent sugar dehydrogenase, ... | | 著者 | Takeda, K, Ishida, T, Yoshida, M, Samejima, M, Ohno, H, Igarashi, K, Nakamura, N. | | 登録日 | 2019-04-09 | | 公開日 | 2019-11-06 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal Structure of the Catalytic and CytochromebDomains in a Eukaryotic Pyrroloquinoline Quinone-Dependent Dehydrogenase.
Appl.Environ.Microbiol., 85, 2019
|
|
6JT6
 
 | | Crystal structure of cytochrome b domain of Pyranose Dehydrogenase from Coprinopsis cinerea | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | 著者 | Takeda, K, Ishida, T, Yoshida, M, Samejima, M, Ohno, H, Igarashi, K, Nakamura, N. | | 登録日 | 2019-04-09 | | 公開日 | 2019-11-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal Structure of the Catalytic and CytochromebDomains in a Eukaryotic Pyrroloquinoline Quinone-Dependent Dehydrogenase.
Appl.Environ.Microbiol., 85, 2019
|
|
3MM2
 
 | | Dye-decolorizing peroxidase (DyP) in complex with cyanide | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYANIDE ION, DyP, ... | | 著者 | Sugano, Y, Yoshida, T, Tsuge, H. | | 登録日 | 2010-04-19 | | 公開日 | 2011-04-27 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | The catalytic mechanism of dye-decolorizing peroxidase DyP may require the swinging movement of an aspartic acid residue
Febs J., 278, 2011
|
|
6K3D
 
 | | Structure of multicopper oxidase mutant | | 分子名称: | COPPER (II) ION, CU-O-CU LINKAGE, Multicopper oxidase | | 著者 | Sakuraba, H, Ohshida, T, Satomura, T, Yoneda, K, Ohshima, T. | | 登録日 | 2019-05-17 | | 公開日 | 2020-05-20 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.919 Å) | | 主引用文献 | Activity enhancement of multicopper oxidase from a hyperthermophile via directed evolution, and its application as the element of a high performance biocathode.
J.Biotechnol., 325, 2021
|
|
1IY6
 
 | | Solution structure of OMSVP3 variant, P14C/N39C | | 分子名称: | OMSVP3 | | 著者 | Hemmi, H, Kumazaki, T, Yamazaki, T, Kojima, S, Yoshida, T, Kyogoku, Y, Katsu, M, Yokosawa, H, Miura, K, Kobayashi, Y. | | 登録日 | 2002-07-23 | | 公開日 | 2003-03-11 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Inhibitory Specificity Change of Ovomucoid Third Domain of the Silver Pheasant upon Introduction of an Engineered Cys14-Cys39 Bond
BIOCHEMISTRY, 42, 2003
|
|
1J1I
 
 | | Crystal structure of a His-tagged Serine Hydrolase Involved in the Carbazole Degradation (CarC enzyme) | | 分子名称: | meta cleavage compound hydrolase | | 著者 | Habe, H, Morii, K, Fushinobu, S, Nam, J.W, Ayabe, Y, Yoshida, T, Wakagi, T, Yamane, H, Nojiri, H, Omori, T. | | 登録日 | 2002-12-05 | | 公開日 | 2003-06-17 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Crystal structure of a histidine-tagged serine hydrolase involved in the carbazole degradation (CarC enzyme).
Biochem.Biophys.Res.Commun., 303, 2003
|
|
1ITO
 
 | | Crystal Structure Analysis of Bovine Spleen Cathepsin B-E64c complex | | 分子名称: | Cathepsin B, N-[1-HYDROXYCARBOXYETHYL-CARBONYL]LEUCYLAMINO-2-METHYL-BUTANE | | 著者 | Yamamoto, A, Tomoo, T, Matsugi, K, Hara, T, In, Y, Murata, M, Kitamura, K, Ishida, T. | | 登録日 | 2002-01-19 | | 公開日 | 2003-01-19 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.286 Å) | | 主引用文献 | Structural basis for development of cathepsin B-specific noncovalent-type inhibitor: crystal structure of cathepsin B-E64c complex
BIOCHIM.BIOPHYS.ACTA, 1597, 2002
|
|
