5LJB
 
 | | Crystal structure of holo human CRBP1 | | Descriptor: | RETINOL, Retinol-binding protein 1 | | Authors: | zanotti, G, Valese, F, Berni, R, menozzi, I. | | Deposit date: | 2016-07-18 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.263 Å) | | Cite: | Structural and molecular determinants affecting the interaction of retinol with human CRBP1.
J. Struct. Biol., 197, 2017
|
|
5LJG
 
 | | Crystal structure of holo human CRBP1 | | Descriptor: | PALMITIC ACID, Retinol-binding protein 1 | | Authors: | Zanotti, G, Vallese, F, Berni, R, Menozzi, I. | | Deposit date: | 2016-07-18 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.146 Å) | | Cite: | Structural and molecular determinants affecting the interaction of retinol with human CRBP1.
J. Struct. Biol., 197, 2017
|
|
5M2X
 
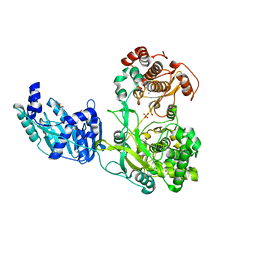 | | Crystal structure of the full-length Zika virus NS5 protein (Human isolate Z1106033) | | Descriptor: | NS5, PHOSPHATE ION, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Ferrero, D.S, Ruiz-Arroyo, V.M, Soler, N, Uson, I, Verdaguer, N. | | Deposit date: | 2016-10-13 | | Release date: | 2018-06-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (4.991 Å) | | Cite: | Supramolecular arrangement of the full-length Zika virus NS5.
Plos Pathog., 15, 2019
|
|
5M15
 
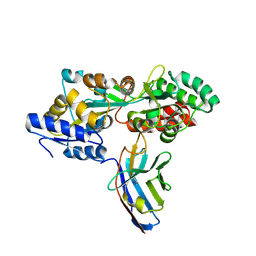 | | Synthetic nanobody in complex with MBP | | Descriptor: | Maltose-binding periplasmic protein, Synthetic nanobody L2_D09, (a-MBP#3) | | Authors: | Zimmermann, I, Egloff, P, Seeger, M.A. | | Deposit date: | 2016-10-07 | | Release date: | 2017-11-15 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synthetic single domain antibodies for the conformational trapping of membrane proteins.
Elife, 7, 2018
|
|
5LXG
 
 | | Revised crystal structure of the human adiponectin receptor 1 in an open conformation | | Descriptor: | Adiponectin receptor protein 1, SULFATE ION, V REGION HEAVY CHAIN, ... | | Authors: | Leyrat, C, Vasiliauskaite-Brooks, I, Granier, S. | | Deposit date: | 2016-09-21 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Structural insights into adiponectin receptors suggest ceramidase activity.
Nature, 6, 2017
|
|
5LJK
 
 | | Crystal structure of human apo CRBP1 | | Descriptor: | Retinol-binding protein 1, SODIUM ION | | Authors: | Zanotti, G, Vallese, F, Berni, R, Menozzi, I. | | Deposit date: | 2016-07-18 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and molecular determinants affecting the interaction of retinol with human CRBP1.
J. Struct. Biol., 197, 2017
|
|
5LXQ
 
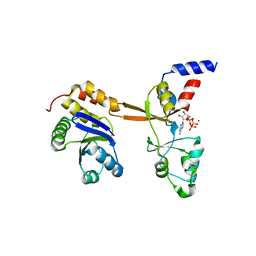 | | Structure of PRL-1 in complex with the Bateman domain of CNNM2 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Metal transporter CNNM2, Protein tyrosine phosphatase type IVA 1, ... | | Authors: | GIMENEZ-Mascarell, P, Oyenarte, I, Hardy, S, Breiderhoff, T, Stuiver, M, Kostantin, E, Diercks, T, Pey, A.L, Ereno-ORBEA, J, Martinez-Chantar, M.L, Khalaf-Nazzal, R, Claverie-Martin, F, Muller, D, Tremblay, M, Martinez-Cruz, L.A. | | Deposit date: | 2016-09-22 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.335 Å) | | Cite: | Structural Basis of the Oncogenic Interaction of Phosphatase PRL-1 with the Magnesium Transporter CNNM2.
J. Biol. Chem., 292, 2017
|
|
5M13
 
 | | Synthetic nanobody in complex with MBP | | Descriptor: | 1,2-ETHANEDIOL, Maltose-binding periplasmic protein, synthetic Nanobody L2_C06 (a-MBP#2) | | Authors: | Zimmermann, I, Egloff, P, Seeger, M.A. | | Deposit date: | 2016-10-07 | | Release date: | 2017-11-15 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.372 Å) | | Cite: | Synthetic single domain antibodies for the conformational trapping of membrane proteins.
Elife, 7, 2018
|
|
5LSR
 
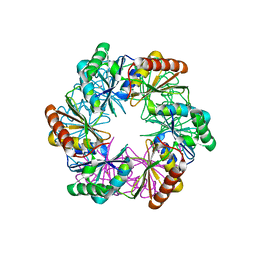 | | Carboxysome shell protein CcmP from Synechococcus elongatus PCC 7942 | | Descriptor: | CcmP, THIOCYANATE ION | | Authors: | Larsson, A.M, Hasse, D, Valegard, K, Andersson, I. | | Deposit date: | 2016-09-05 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of beta-carboxysome shell protein CcmP: ligand binding correlates with the closed or open central pore.
J. Exp. Bot., 68, 2017
|
|
5LY7
 
 | | Crystal structure of NagZ H174A mutant from Pseudomonas aeruginosa in complex with the inhibitor 2-acetamido-1,2-dideoxynojirimycin | | Descriptor: | 2-ACETAMIDO-1,2-DIDEOXYNOJIRMYCIN, Beta-hexosaminidase, DI(HYDROXYETHYL)ETHER | | Authors: | Acebron, I, Artola-Recolons, C, Mahasenan, K, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2016-09-25 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Catalytic Cycle of the N-Acetylglucosaminidase NagZ from Pseudomonas aeruginosa.
J. Am. Chem. Soc., 139, 2017
|
|
6MGS
 
 | | Crystal structure of alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde-Decarboxylase with Space Group of C2221 | | Descriptor: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, COBALT (II) ION | | Authors: | Yang, Y, Davis, I, Matsui, T, Rubalcava, I, Liu, A. | | Deposit date: | 2018-09-14 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.131 Å) | | Cite: | Quaternary structure of alpha-amino-beta-carboxymuconate-ε-semialdehyde decarboxylase (ACMSD) controls its activity.
J.Biol.Chem., 294, 2019
|
|
6OCP
 
 | | Crystal structure of a human GABAB receptor peptide bound to KCTD16 T1 | | Descriptor: | BTB/POZ domain-containing protein KCTD16, Gamma-aminobutyric acid type B receptor subunit 2 | | Authors: | Zuo, H, Glaaser, I, Zhao, Y, Kurinov, I, Mosyak, L, Wang, H, Liu, J, Park, J, Frangaj, A, Sturchler, E, Zhou, M, McDonald, P, Geng, Y, Slesinger, P.A, Fan, Q.R. | | Deposit date: | 2019-03-25 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for auxiliary subunit KCTD16 regulation of the GABABreceptor.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4WXB
 
 | | Crystal Structure of Serine Hydroxymethyltransferase from Streptococcus thermophilus | | Descriptor: | CACODYLATE ION, CITRIC ACID, GLYCEROL, ... | | Authors: | Hernandez, K, Zelen, I, Petrillo, G, Uson, I, Wandtke, C, Bujons, J, Joglar, J, Parella, T, Clapes, P. | | Deposit date: | 2014-11-13 | | Release date: | 2015-02-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Engineered L-Serine Hydroxymethyltransferase from Streptococcus thermophilus for the Synthesis of alpha , alpha-Dialkyl-alpha-Amino Acids.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4WJS
 
 | |
4WJV
 
 | | Crystal structure of Rsa4 in complex with the Nsa2 binding peptide | | Descriptor: | Maltose-binding periplasmic protein, Ribosome assembly protein 4, Ribosome biogenesis protein NSA2, ... | | Authors: | Holdermann, I, Paternoga, H, Bassler, J, Hurt, E, Sinning, I. | | Deposit date: | 2014-10-01 | | Release date: | 2014-11-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A network of assembly factors is involved in remodeling rRNA elements during preribosome maturation.
J.Cell Biol., 207, 2014
|
|
4WJU
 
 | | Crystal structure of Rsa4 from Saccharomyces cerevisiae | | Descriptor: | GLYCEROL, Ribosome assembly protein 4 | | Authors: | Holdermann, I, Bassler, J, Hurt, E, Sinning, I. | | Deposit date: | 2014-10-01 | | Release date: | 2014-11-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A network of assembly factors is involved in remodeling rRNA elements during preribosome maturation.
J.Cell Biol., 207, 2014
|
|
5DWX
 
 | |
9ESM
 
 | |
5E31
 
 | | 2.3 Angstrom Crystal Structure of the Monomeric Form of Penicillin Binding Protein 2 Prime from Enterococcus faecium. | | Descriptor: | Penicillin binding protein 2 prime | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Dubrovska, I, Flores, K, Filippova, E, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-10-01 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 2.3 Angstrom Crystal Structure of the Monomeric Form of Penicillin Binding Protein 2 Prime from Enterococcus faecium.
To Be Published
|
|
5E3X
 
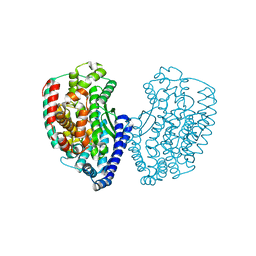 | | Crystal structure of thermostable Carboxypeptidase (FisCP) from Fervidobacterium Islandicum AW-1 | | Descriptor: | COBALT (II) ION, Thermostable carboxypeptidase 1 | | Authors: | Dhanasingh, I, Lee, Y.-J, Lee, D.W, Lee, S.H. | | Deposit date: | 2015-10-05 | | Release date: | 2016-02-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | Biochemical and structural characterization of a keratin-degrading M32 carboxypeptidase from Fervidobacterium islandicum AW-1
Biochem.Biophys.Res.Commun., 468, 2015
|
|
5E8P
 
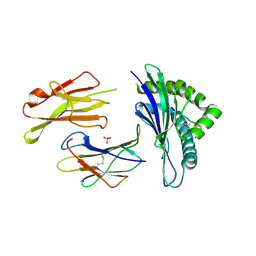 | | The structure of the TEIPP associated altered peptide ligand Trh4-p5NLE in complex with H-2D(b) | | Descriptor: | Beta-2-microglobulin, Ceramide synthase 5, H-2 class I histocompatibility antigen, ... | | Authors: | Hafstrand, I, Doorduijn, E, Duru, A.D, Buratto, J, Oliveira, C.C, Sandalova, T, van Hall, T, Achour, A. | | Deposit date: | 2015-10-14 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The MHC Class I Cancer-Associated Neoepitope Trh4 Linked with Impaired Peptide Processing Induces a Unique Noncanonical TCR Conformer.
J Immunol., 196, 2016
|
|
5EBH
 
 | |
5DVY
 
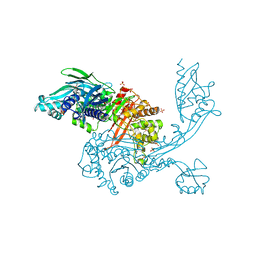 | | 2.95 Angstrom Crystal Structure of the Dimeric Form of Penicillin Binding Protein 2 Prime from Enterococcus faecium | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Penicillin binding protein 2 prime, SULFATE ION | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Dubrovska, I, Flores, K, Filippova, E, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-09-21 | | Release date: | 2015-10-07 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | 2.95 Angstrom Crystal Structure of the Dimeric Form of Penicillin Binding Protein 2 Prime from Enterococcus faecium.
To Be Published
|
|
5ECT
 
 | | Mycobacterium tuberculosis dUTPase G143STOP mutant | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Nagy, G.N, Leveles, I, Harmat, V, Vertessy, G.B. | | Deposit date: | 2015-10-20 | | Release date: | 2016-11-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural Characterization of Arginine Fingers: Identification of an Arginine Finger for the Pyrophosphatase dUTPases.
J. Am. Chem. Soc., 138, 2016
|
|
5E4W
 
 | | Crystal structure of cpSRP43 chromodomains 2 and 3 in complex with the Alb3 tail | | Descriptor: | CALCIUM ION, GLYCEROL, Inner membrane protein ALBINO3, ... | | Authors: | Horn, A, Ahmed, Y.L, Wild, K, Sinning, I. | | Deposit date: | 2015-10-07 | | Release date: | 2015-12-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for cpSRP43 chromodomain selectivity and dynamics in Alb3 insertase interaction.
Nat Commun, 6, 2015
|
|
