8DIB
 
 | |
8DIG
 
 | |
8DIH
 
 | | Virtual screening for novel SARS-CoV-2 main protease non-covalent and covalent inhibitors | | 分子名称: | (1P,1'R)-1-(isoquinolin-4-yl)-2',3'-dihydrospiro[imidazolidine-4,1'-indene]-2,5-dione, 3C-like proteinase nsp5 | | 著者 | Singh, I, Shoichet, B.K. | | 登録日 | 2022-06-29 | | 公開日 | 2023-06-28 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | Large library docking for novel SARS-CoV-2 main protease non-covalent and covalent inhibitors.
Protein Sci., 32, 2023
|
|
8DIF
 
 | |
8DIE
 
 | |
8DII
 
 | |
7ABU
 
 | | Structure of SARS-CoV-2 Main Protease bound to RS102895 | | 分子名称: | 1'-[2-[4-(trifluoromethyl)phenyl]ethyl]spiro[1~{H}-3,1-benzoxazine-4,4'-piperidine]-2-one, 3C-like proteinase, DIMETHYL SULFOXIDE, ... | | 著者 | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Lane, T.J, Dunkel, I, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | 登録日 | 2020-09-08 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7ADW
 
 | | Structure of SARS-CoV-2 Main Protease bound to 2,4'-Dimethylpropiophenone. | | 分子名称: | 2-methyl-1-(4-methylphenyl)propan-1-one, 3C-like proteinase, CHLORIDE ION, ... | | 著者 | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | 登録日 | 2020-09-16 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7A1U
 
 | | Structure of SARS-CoV-2 Main Protease bound to Fusidic Acid. | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, FUSIDIC ACID, ... | | 著者 | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Andaleeb, H, Werner, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | 登録日 | 2020-08-14 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
8DID
 
 | |
8F41
 
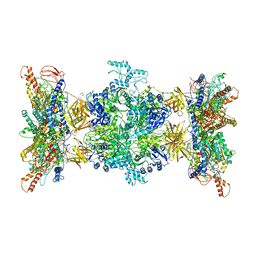 | | 3-methylcrotonyl-CoA carboxylase in filament, alpha-subunit centered | | 分子名称: | 3-methylcrotonyl-CoA carboxylase, alpha-subunit, beta-subunit, ... | | 著者 | Hu, J.J, Lee, J.K.J, Liu, Y.T, Yu, C, Huang, L, Afasizheva, I, Afasizhev, R, Zhou, Z.H. | | 登録日 | 2022-11-10 | | 公開日 | 2023-01-11 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Discovery, structure, and function of filamentous 3-methylcrotonyl-CoA carboxylase.
Structure, 31, 2023
|
|
5YK9
 
 | |
5Y9T
 
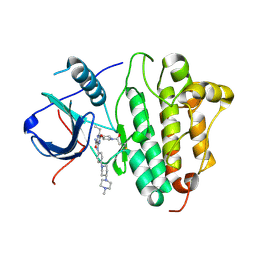 | | Crystal Structure of EGFR T790M mutant in complex with naquotinib | | 分子名称: | 6-ethyl-3-[[4-[4-(4-methylpiperazin-1-yl)piperidin-1-yl]phenyl]amino]-5-[(3R)-1-prop-2-enoylpyrrolidin-3-yl]oxy-pyrazin e-2-carboxamide, Epidermal growth factor receptor | | 著者 | Mimasu, S, Tomimoto, Y, Maiko, I, Yasushi, A, Tatsuya, N. | | 登録日 | 2017-08-28 | | 公開日 | 2018-07-11 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.25 Å) | | 主引用文献 | Pharmacological and Structural Characterizations of Naquotinib, a Novel Third-Generation EGFR Tyrosine Kinase Inhibitor, inEGFR-Mutated Non-Small Cell Lung Cancer.
Mol. Cancer Ther., 17, 2018
|
|
6HKC
 
 | |
6A28
 
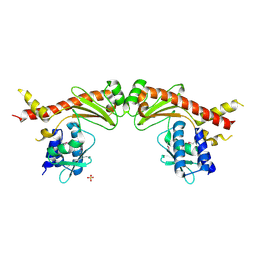 | | Crystal structure of PprA W183R mutant form 2 | | 分子名称: | DNA repair protein PprA, SULFATE ION | | 著者 | Adachi, M, Shibazaki, C, Shimizu, R, Arai, S, Satoh, K, Narumi, I, Kuroki, R. | | 登録日 | 2018-06-09 | | 公開日 | 2018-12-26 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.193 Å) | | 主引用文献 | Extended structure of pleiotropic DNA repair-promoting protein PprA from Deinococcus radiodurans.
FASEB J., 33, 2019
|
|
9G13
 
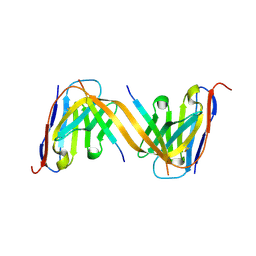 | | VHH H3-2 in complex with Tau C-terminal peptide | | 分子名称: | Isoform Tau-F of Microtubule-associated protein tau, VHH H3-2 | | 著者 | Dupre, E, Landrieu, I, Danis, C, Hanoulle, X, Mortelecque, J. | | 登録日 | 2024-07-09 | | 公開日 | 2024-07-31 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | VHH H3-2 in complex with Tau C-terminal peptide
To Be Published
|
|
1A2S
 
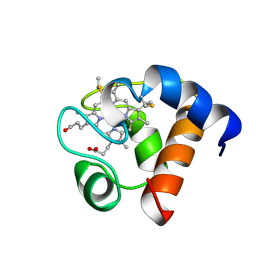 | | THE SOLUTION NMR STRUCTURE OF OXIDIZED CYTOCHROME C6 FROM THE GREEN ALGA MONORAPHIDIUM BRAUNII, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | CYTOCHROME C6, HEME C | | 著者 | Banci, L, Bertini, I, De La Rosa, M.A, Koulougliotis, D, Navarro, J.A, Walter, O. | | 登録日 | 1998-01-10 | | 公開日 | 1998-04-29 | | 最終更新日 | 2022-02-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of oxidized cytochrome c6 from the green alga Monoraphidium braunii.
Biochemistry, 37, 1998
|
|
4WXF
 
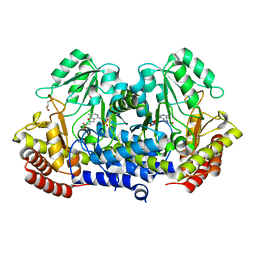 | | Crystal structure of L-Serine Hydroxymethyltransferase in complex with glycine | | 分子名称: | GLYCEROL, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], Serine hydroxymethyltransferase | | 著者 | Hernandez, K, Zelen, I, Petrillo, G, Uson, I, Wadtke, C, Bujons, J, Joglar, J, Parella, T, Clapes, P. | | 登録日 | 2014-11-13 | | 公開日 | 2015-02-04 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Engineered L-Serine Hydroxymethyltransferase from Streptococcus thermophilus for the Synthesis of alpha , alpha-Dialkyl-alpha-Amino Acids.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
121D
 
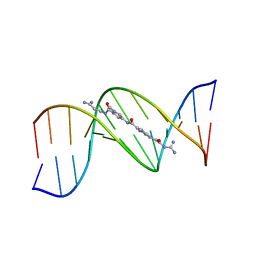 | | MOLECULAR STRUCTURE OF THE A-TRACT DNA DODECAMER D(CGCAAATTTGCG) COMPLEXED WITH THE MINOR GROOVE BINDING DRUG NETROPSIN | | 分子名称: | DNA (5'-D(*CP*GP*CP*AP*AP*AP*TP*TP*TP*GP*CP*G)-3'), NETROPSIN | | 著者 | Tabernero, L, Verdaguer, N, Coll, M, Fita, I, Van Der Marel, G.A, Van Boom, J.H, Rich, A, Aymami, J. | | 登録日 | 1993-04-14 | | 公開日 | 1994-01-15 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Molecular structure of the A-tract DNA dodecamer d(CGCAAATTTGCG) complexed with the minor groove binding drug netropsin.
Biochemistry, 32, 1993
|
|
132L
 
 | |
1AK8
 
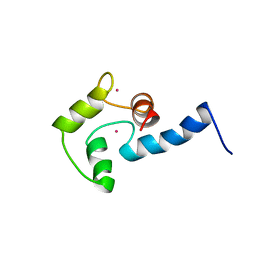 | | NMR SOLUTION STRUCTURE OF CERIUM-LOADED CALMODULIN AMINO-TERMINAL DOMAIN (CE2-TR1C), 23 STRUCTURES | | 分子名称: | CALMODULIN, CERIUM (III) ION | | 著者 | Bentrop, D, Bertini, I, Cremonini, M.A, Forsen, S, Luchinat, C, Malmendal, A. | | 登録日 | 1997-05-29 | | 公開日 | 1997-09-17 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the paramagnetic complex of the N-terminal domain of calmodulin with two Ce3+ ions by 1H NMR.
Biochemistry, 36, 1997
|
|
1A49
 
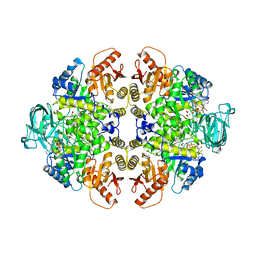 | | BIS MG-ATP-K-OXALATE COMPLEX OF PYRUVATE KINASE | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, OXALATE ION, ... | | 著者 | Larsen, T.M, Benning, M.M, Rayment, I, Reed, G.H. | | 登録日 | 1998-02-12 | | 公開日 | 1999-04-20 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure of the bis(Mg2+)-ATP-oxalate complex of the rabbit muscle pyruvate kinase at 2.1 A resolution: ATP binding over a barrel.
Biochemistry, 37, 1998
|
|
1AD5
 
 | |
1AMQ
 
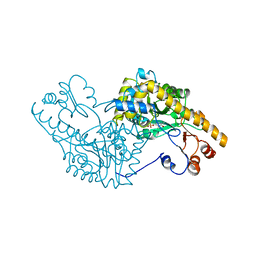 | | X-RAY CRYSTALLOGRAPHIC STUDY OF PYRIDOXAMINE 5'-PHOSPHATE-TYPE ASPARTATE AMINOTRANSFERASES FROM ESCHERICHIA COLI IN THREE FORMS | | 分子名称: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ASPARTATE AMINOTRANSFERASE | | 著者 | Miyahara, I, Hirotsu, K, Hayashi, H, Kagamiyama, H. | | 登録日 | 1994-07-01 | | 公開日 | 1994-09-30 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | X-ray crystallographic study of pyridoxamine 5'-phosphate-type aspartate aminotransferases from Escherichia coli in three forms.
J.Biochem.(Tokyo), 116, 1994
|
|
1AO2
 
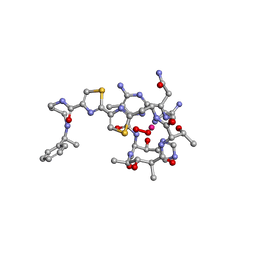 | | cobalt(III)-deglycopepleomycin determined by NMR studies | | 分子名称: | AGLYCON OF PEPLOMYCIN, COBALT (III) ION, HYDROGEN PEROXIDE | | 著者 | Caceres-Cortes, J, Sugiyama, H, Ikudome, K, Saito, I, Wang, A.H.-J. | | 登録日 | 1997-07-16 | | 公開日 | 1999-07-30 | | 最終更新日 | 2024-03-13 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structures of cobalt(III)-pepleomycin and cobalt(III)-deglycopepleomycin (green forms) determined by NMR studies.
Eur.J.Biochem., 244, 1997
|
|
