5S4I
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with SF051 | | Descriptor: | (5S)-1-(4-chlorophenyl)-5-methylimidazolidine-2,4-dione, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.131 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
2EV4
 
 | | Structure of Rv1264N, the regulatory domain of the mycobacterial adenylyl cylcase Rv1264, with a salt precipitant | | Descriptor: | CHLORIDE ION, Hypothetical protein Rv1264/MT1302, OLEIC ACID | | Authors: | Findeisen, F, Tews, I, Sinning, I. | | Deposit date: | 2005-10-30 | | Release date: | 2006-11-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | The structure of the regulatory domain of the adenylyl cyclase Rv1264 from Mycobacterium tuberculosis with bound oleic acid
J.Mol.Biol., 369, 2007
|
|
5S4H
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with SF048 | | Descriptor: | 1-carbamoylpiperidine-4-carboxylic acid, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.175 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S4F
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with SF003 | | Descriptor: | 1,8-naphthyridine, Non-structural protein 3, SULFATE ION | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.131 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S4G
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with SF005 | | Descriptor: | Non-structural protein 3, SULFATE ION, [1,2,4]triazolo[4,3-a]pyridin-3-amine | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.172 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
2CJB
 
 | | Crystal structure of Methanosarcina barkeri seryl-tRNA synthetase complexed with serine | | Descriptor: | CHLORIDE ION, SERINE, SERYL-TRNA SYNTHETASE, ... | | Authors: | Bilokapic, S, Maier, T, Ahel, D, Gruic-Sovulj, I, Soll, D, Weygand-Durasevic, I, Ban, N. | | Deposit date: | 2006-03-30 | | Release date: | 2006-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the Unusual Seryl-tRNA Synthetase Reveals a Distinct Zinc-Dependent Mode of Substrate Recognition
Embo J., 25, 2006
|
|
2D31
 
 | | Crystal structure of disulfide-linked HLA-G dimer | | Descriptor: | 9-mer peptide from Histone H2A, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Shiroishi, M, Kuroki, K, Ose, T, Rasubala, L, Shiratori, I, Arase, H, Tsumoto, K, Kumagai, I, Kohda, D, Maenaka, K. | | Deposit date: | 2005-09-23 | | Release date: | 2006-03-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Efficient Leukocyte Ig-like Receptor Signaling and Crystal Structure of Disulfide-linked HLA-G Dimer
J.Biol.Chem., 281, 2006
|
|
6HPN
 
 | | Crystal structure of Borrelia spielmanii WP_012665240 (BSA64) - an orthologous protein to B. burgdorferi BBA64 | | Descriptor: | Antigen, P35 | | Authors: | Brangulis, K, Akopjana, I, Kazaks, A, Tars, K. | | Deposit date: | 2018-09-21 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural analysis of the outer surface proteins from Borrelia burgdorferi paralogous gene family 54 that are thought to be the key players in the pathogenesis of Lyme disease.
J.Struct.Biol., 210, 2020
|
|
2WK1
 
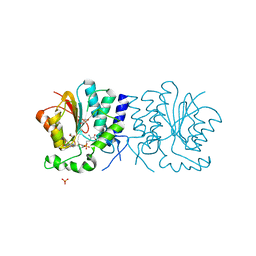 | | Structure of the O-methyltransferase NovP | | Descriptor: | 1,2-ETHANEDIOL, NOVP, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Gomez Garcia, I, Stevenson, C.E.M, Uson, I, Freel Meyers, C.L, Walsh, C.T, Lawson, D.M. | | Deposit date: | 2009-06-03 | | Release date: | 2009-12-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Crystal Structure of the Novobiocin Biosynthetic Enzyme Novp: The First Representative Structure for the Tylf O-Methyltransferase Superfamily.
J.Mol.Biol., 395, 2010
|
|
5M93
 
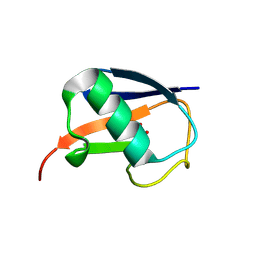 | |
5MU7
 
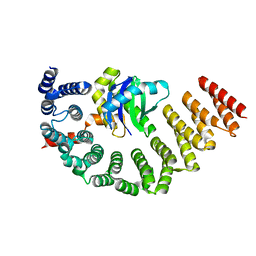 | | Crystal Structure of the beta/delta-COPI Core Complex | | Descriptor: | Coatomer subunit beta, Coatomer subunit delta-like protein | | Authors: | Kopp, J, Aderhold, P, Wieland, F, Sinning, I. | | Deposit date: | 2017-01-12 | | Release date: | 2017-06-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | 9 angstrom structure of the COPI coat reveals that the Arf1 GTPase occupies two contrasting molecular environments.
Elife, 6, 2017
|
|
7OTE
 
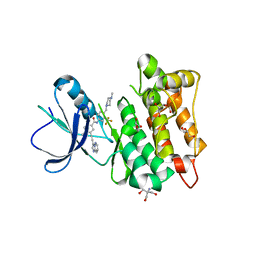 | | Src Kinase Domain in complex with ponatinib | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-(imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzam ide, GLYCEROL, ... | | Authors: | Soriano-Maldonado, P, Cuesta-Hernandez, H.N, Plaza-Menacho, I. | | Deposit date: | 2021-06-10 | | Release date: | 2022-06-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | An allosteric switch between the activation loop and a c-terminal palindromic phospho-motif controls c-Src function.
Nat Commun, 14, 2023
|
|
2XIM
 
 | | ARGININE RESIDUES AS STABILIZING ELEMENTS IN PROTEINS | | Descriptor: | D-XYLOSE ISOMERASE, MAGNESIUM ION, Xylitol | | Authors: | Mrabet, N.T, Van Denbroek, A, Van Den Brande, I, Stanssens, P, Laroche, Y, Lambeir, A.-M, Matthyssens, G, Jenkins, J, Chiadmi, M, Vantilbeurgh, H, Rey, F, Janin, J, Quax, W.J, Lasters, I, Demaeyer, M, Wodak, S.J. | | Deposit date: | 1991-05-29 | | Release date: | 1993-04-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Arginine residues as stabilizing elements in proteins.
Biochemistry, 31, 1992
|
|
5MU6
 
 | | Human N-myristoyltransferase (NMT1) with Myristoyl-CoA and IMP-1088 inhibitor bound | | Descriptor: | 1-[5-[3,4-bis(fluoranyl)-2-[2-(1,3,5-trimethylpyrazol-4-yl)ethoxy]phenyl]-1-methyl-indazol-3-yl]-~{N},~{N}-dimethyl-methanamine, GLYCEROL, Glycylpeptide N-tetradecanoyltransferase 1, ... | | Authors: | Perez-Dorado, I, Bell, A.S, Tate, E.W. | | Deposit date: | 2017-01-12 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Fragment-derived inhibitors of human N-myristoyltransferase block capsid assembly and replication of the common cold virus.
Nat Chem, 10, 2018
|
|
1N9C
 
 | |
5NDX
 
 | | The bacterial orthologue of Human a-L-iduronidase does not need N-glycan post-translational modifications to be catalytically competent: Crystallography and QM/MM insights into Mucopolysaccharidosis I | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{S})-6-(4-methyl-2-oxidanylidene-chromen-7-yl)oxy-3,4,5-tris(oxidanyl)oxane-2-carboxylic acid, Glycosyl hydrolase, SULFATE ION | | Authors: | Raich, L, Valero-Gonzalez, J, Castro-Lopez, J, Millan, C, Jimenez-Garcia, M.J, Nieto, P, Uson, I, Hurtado-Guerrero, R, Rovira, C. | | Deposit date: | 2017-03-09 | | Release date: | 2018-07-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The bacterial orthologue of Human a-L-iduronidase does not need N-glycan post-translational modifications to be catalytically competent:
Crystallography and QM/MM insights into Mucopolysaccharidosis I.
To Be Published
|
|
5NCW
 
 | | Structure of the trypsin induced serpin-type proteinase inhibitor, miropin (V367K/K368A mutant). | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Goulas, T, Ksiazek, M, Garcia-Ferrer, I, Mizgalska, D, Potempa, J, Gomis-Ruth, X. | | Deposit date: | 2017-03-06 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A structure-derived snap-trap mechanism of a multispecific serpin from the dysbiotic human oral microbiome.
J. Biol. Chem., 292, 2017
|
|
3Q0Y
 
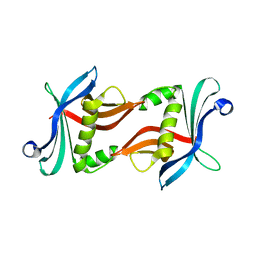 | | N-terminal domain of C. reinhardtii SAS-6 homolog Bld12p | | Descriptor: | Centriole protein | | Authors: | Kitagawa, D, Vakonakis, I, Olieric, N, Hilbert, M, Keller, D, Olieric, V, Bortfeld, M, Erat, M.C, Flueckiger, I, Goenczy, P, Steinmetz, M.O. | | Deposit date: | 2010-12-16 | | Release date: | 2011-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of the 9-fold symmetry of centrioles.
Cell(Cambridge,Mass.), 144, 2011
|
|
3Q0X
 
 | | N-terminal coiled-coil dimer domain of C. reinhardtii SAS-6 homolog Bld12p | | Descriptor: | Centriole protein | | Authors: | Kitagawa, D, Vakonakis, I, Olieric, N, Hilbert, M, Keller, D, Olieric, V, Bortfeld, M, Erat, M.C, Flueckiger, I, Goenczy, P, Steinmetz, M.O. | | Deposit date: | 2010-12-16 | | Release date: | 2011-02-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Structural basis of the 9-fold symmetry of centrioles.
Cell(Cambridge,Mass.), 144, 2011
|
|
5NCS
 
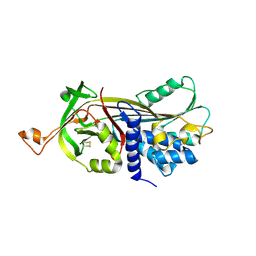 | | Structure of the native serpin-type proteinase inhibitor, miropin. | | Descriptor: | Serpin | | Authors: | Goulas, T, Ksiazek, M, Garcia-Ferrer, I, Mizgalska, D, Potempa, J, Gomis-Ruth, X. | | Deposit date: | 2017-03-06 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A structure-derived snap-trap mechanism of a multispecific serpin from the dysbiotic human oral microbiome.
J. Biol. Chem., 292, 2017
|
|
5NCU
 
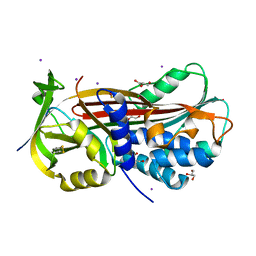 | | Structure of the subtilisin induced serpin-type proteinase inhibitor, miropin. | | Descriptor: | CHLORIDE ION, GLYCEROL, IODIDE ION, ... | | Authors: | Goulas, T, Ksiazek, M, Garcia-Ferrer, I, Mizgalska, D, Potempa, J, Gomis-Ruth, X. | | Deposit date: | 2017-03-06 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A structure-derived snap-trap mechanism of a multispecific serpin from the dysbiotic human oral microbiome.
J. Biol. Chem., 292, 2017
|
|
5NCT
 
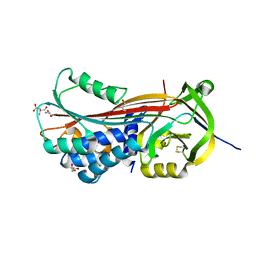 | | Structure of the trypsin induced serpin-type proteinase inhibitor, miropin. | | Descriptor: | ASPARTIC ACID, GLYCEROL, SERINE, ... | | Authors: | Goulas, T, Ksiazek, M, Garcia-Ferrer, I, Mizgalska, D, Potempa, J, Gomis-Ruth, X. | | Deposit date: | 2017-03-06 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A structure-derived snap-trap mechanism of a multispecific serpin from the dysbiotic human oral microbiome.
J. Biol. Chem., 292, 2017
|
|
6J82
 
 | | Crystal structure of TleB apo | | Descriptor: | Cytochrome P-450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Alblova, M, Nakamura, H, Mori, T, Abe, I. | | Deposit date: | 2019-01-18 | | Release date: | 2019-08-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Molecular basis for the P450-catalyzed C-N bond formation in indolactam biosynthesis.
Nat.Chem.Biol., 15, 2019
|
|
6J88
 
 | | Crystal structure of HinD with benzo[b]thiophen analog | | Descriptor: | N-[(2S)-1-(1-benzothiophen-3-yl)-3-hydroxypropan-2-yl]-N~2~-methyl-L-valinamide, Nocardicin N-oxygenase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Fei, H, Mori, T, Abe, I. | | Deposit date: | 2019-01-18 | | Release date: | 2019-08-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular basis for the P450-catalyzed C-N bond formation in indolactam biosynthesis.
Nat.Chem.Biol., 15, 2019
|
|
7NNP
 
 | | Rb-loaded cryo-EM structure of the E1-ATP KdpFABC complex. | | Descriptor: | CARDIOLIPIN, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Potassium-transporting ATPase ATP-binding subunit, ... | | Authors: | Silberberg, J.M, Corey, R.A, Hielkema, L, Stock, C, Stansfeld, P.J, Paulino, C, Haenelt, I. | | Deposit date: | 2021-02-25 | | Release date: | 2021-07-28 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Deciphering ion transport and ATPase coupling in the intersubunit tunnel of KdpFABC.
Nat Commun, 12, 2021
|
|
