6YFD
 
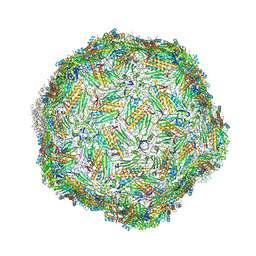 | | Virus-like particle of Beihai levi-like virus 14 | | Descriptor: | coat protein | | Authors: | Rumnieks, J, Kalnins, G, Sisovs, M, Lieknina, I, Tars, K. | | Deposit date: | 2020-03-26 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Three-dimensional structure of 22 uncultured ssRNA bacteriophages: Flexibility of the coat protein fold and variations in particle shapes.
Sci Adv, 6, 2020
|
|
6YFS
 
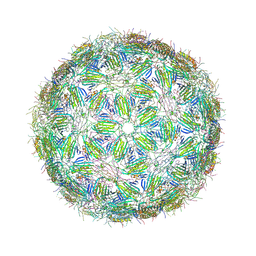 | | Virus-like particle of bacteriophage PQ-465 | | Descriptor: | coat protein | | Authors: | Rumnieks, J, Kalnins, G, Sisovs, M, Lieknina, I, Tars, K. | | Deposit date: | 2020-03-26 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.498 Å) | | Cite: | Three-dimensional structure of 22 uncultured ssRNA bacteriophages: Flexibility of the coat protein fold and variations in particle shapes.
Sci Adv, 6, 2020
|
|
6YF9
 
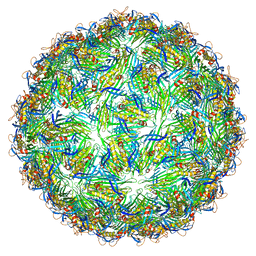 | | Virus-like particle of bacteriophage AVE002 | | Descriptor: | coat protein | | Authors: | Rumnieks, J, Kalnins, G, Sisovs, M, Lieknina, I, Tars, K. | | Deposit date: | 2020-03-26 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.196 Å) | | Cite: | Three-dimensional structure of 22 uncultured ssRNA bacteriophages: Flexibility of the coat protein fold and variations in particle shapes.
Sci Adv, 6, 2020
|
|
8GKF
 
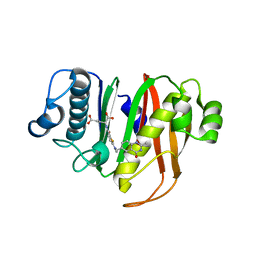 | |
1VAD
 
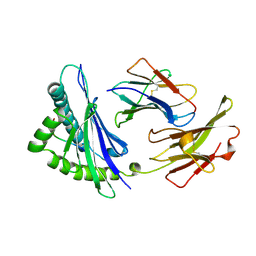 | |
5I3C
 
 | | Crystal structure of E.coli purine nucleoside phosphorylase with acycloguanosine | | Descriptor: | 9-HYROXYETHOXYMETHYLGUANINE, Purine nucleoside phosphorylase DeoD-type, SULFATE ION | | Authors: | Timofeev, V.I, Abramchik, Y.A, Esipov, R.S, Kuranova, I.P. | | Deposit date: | 2016-02-10 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Crystal structure of Escherichia coli purine nucleoside phosphorylase complexed with acyclovir.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
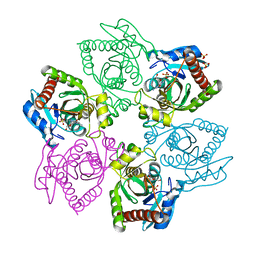
5MVV
 
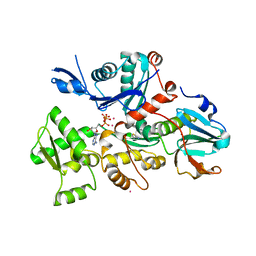 | | Crystal structure of Plasmodium falciparum actin I- gelsolin segment 1 -CdATP complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-1, CADMIUM ION, ... | | Authors: | Panneerselvam, S, Kumpula, E.-P, Kursula, I, Burkhardt, A, Meents, A. | | Deposit date: | 2017-01-17 | | Release date: | 2017-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Rapid cadmium SAD phasing at the standard wavelength (1 angstrom ).
Acta Crystallogr D Struct Biol, 73, 2017
|
|
6YFC
 
 | | Virus-like particle of bacteriophage AVE019 | | Descriptor: | CALCIUM ION, coat protein | | Authors: | Rumnieks, J, Kalnins, G, Sisovs, M, Lieknina, I, Tars, K. | | Deposit date: | 2020-03-26 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.246 Å) | | Cite: | Three-dimensional structure of 22 uncultured ssRNA bacteriophages: Flexibility of the coat protein fold and variations in particle shapes.
Sci Adv, 6, 2020
|
|
8K9L
 
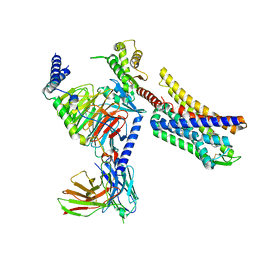 | | Full agonist- and positive allosteric modulator-bound mu-type opioid receptor-G protein complex | | Descriptor: | (2~{S})-2-(3-bromanyl-4-methoxy-phenyl)-3-(4-chlorophenyl)sulfonyl-1,3-thiazolidine, DAMGO, G protein subunit alpha i3, ... | | Authors: | Hisano, T, Uchikubo-Kamo, T, Shirouzu, M, Imai, S, Kaneko, S, Shimada, I. | | Deposit date: | 2023-08-01 | | Release date: | 2024-05-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural and dynamic insights into the activation of the mu-opioid receptor by an allosteric modulator.
Nat Commun, 15, 2024
|
|
5MU6
 
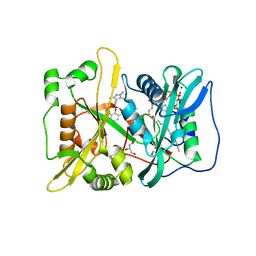 | | Human N-myristoyltransferase (NMT1) with Myristoyl-CoA and IMP-1088 inhibitor bound | | Descriptor: | 1-[5-[3,4-bis(fluoranyl)-2-[2-(1,3,5-trimethylpyrazol-4-yl)ethoxy]phenyl]-1-methyl-indazol-3-yl]-~{N},~{N}-dimethyl-methanamine, GLYCEROL, Glycylpeptide N-tetradecanoyltransferase 1, ... | | Authors: | Perez-Dorado, I, Bell, A.S, Tate, E.W. | | Deposit date: | 2017-01-12 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Fragment-derived inhibitors of human N-myristoyltransferase block capsid assembly and replication of the common cold virus.
Nat Chem, 10, 2018
|
|
7ATF
 
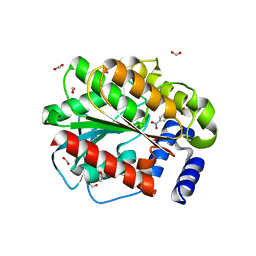 | | Structure of EstD11 in complex with p-Nitrophenol | | Descriptor: | ACETATE ION, EstD11, FORMIC ACID, ... | | Authors: | Miguel-Ruano, V, Rivera, I, Hermoso, J.A. | | Deposit date: | 2020-10-30 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Biochemical and Structural Characterization of a novel thermophilic esterase EstD11 provide catalytic insights for the HSL family.
Comput Struct Biotechnol J, 19, 2021
|
|
7AV5
 
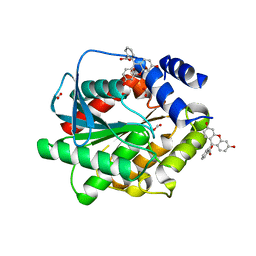 | | Structure of EstD11 in complex with Fluorescein | | Descriptor: | ACETATE ION, EstD11, FLUORESCIN, ... | | Authors: | Miguel-Ruano, V, Rivera, I, Hermoso, J.A. | | Deposit date: | 2020-11-04 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Biochemical and Structural Characterization of a novel thermophilic esterase EstD11 provide catalytic insights for the HSL family.
Comput Struct Biotechnol J, 19, 2021
|
|
7AT3
 
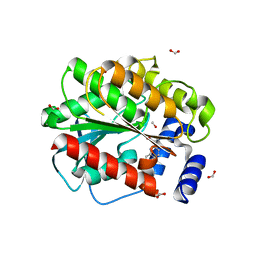 | | Structure of EstD11 in complex with Naproxen and methanol | | Descriptor: | (2R)-2-(6-methoxynaphthalen-2-yl)propanoic acid, EstD11, FORMIC ACID, ... | | Authors: | Miguel-Ruano, V, Rivera, I, Hermoso, J.A. | | Deposit date: | 2020-10-28 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Biochemical and Structural Characterization of a novel thermophilic esterase EstD11 provide catalytic insights for the HSL family.
Comput Struct Biotechnol J, 19, 2021
|
|
7AT0
 
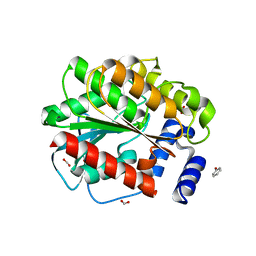 | |
7ATD
 
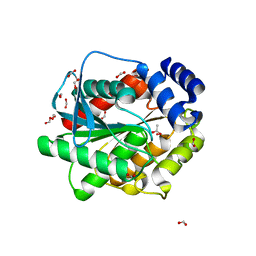 | | Structure of inactive EstD11 S144A in complex with methyl-naproxen | | Descriptor: | ACETATE ION, EstD11 S144A, FORMIC ACID, ... | | Authors: | Miguel-Ruano, V, Rivera, I, Hermoso, J.A. | | Deposit date: | 2020-10-29 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Biochemical and Structural Characterization of a novel thermophilic esterase EstD11 provide catalytic insights for the HSL family.
Comput Struct Biotechnol J, 19, 2021
|
|
7ATQ
 
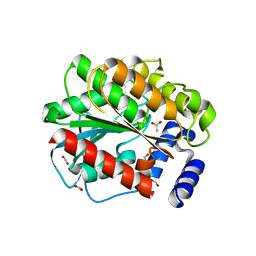 | | Structure of EstD11 in complex with cyclohexane carboxylic acid | | Descriptor: | ACETATE ION, EstD11, FORMIC ACID, ... | | Authors: | Miguel-Ruano, V, Rivera, I, Hermoso, J.A. | | Deposit date: | 2020-10-30 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Biochemical and Structural Characterization of a novel thermophilic esterase EstD11 provide catalytic insights for the HSL family.
Comput Struct Biotechnol J, 19, 2021
|
|
7AUY
 
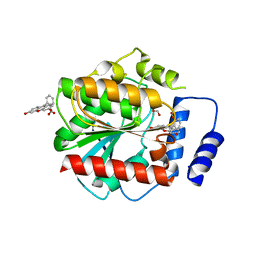 | |
5MS9
 
 | |
6OFK
 
 | | Crystal structure of green fluorescent protein (GFP); S65T; ih circular permutant (50-51) | | Descriptor: | ACETATE ION, Green Fluorescent Protein (GFP); S65T; ih circular permutant (50-51) | | Authors: | Lin, C.-Y, Romei, M.G, Mathews, I.I, Boxer, S.G. | | Deposit date: | 2019-03-30 | | Release date: | 2019-07-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Unified Model for Photophysical and Electro-Optical Properties of Green Fluorescent Proteins.
J.Am.Chem.Soc., 141, 2019
|
|
6GNU
 
 | | Crystal Structure of Leishmania major N-Myristoyltransferase (NMT) With Bound Myristoyl-CoA and a Quionlinyl aryl sulphonamide ligand | | Descriptor: | 4-[4-(1-methylpiperidin-4-yl)butyl]-~{N}-[6-(2-methylpropyl)quinolin-5-yl]benzenesulfonamide, Glycylpeptide N-tetradecanoyltransferase, TETRADECANOYL-COA | | Authors: | Robinson, D.A, Harrison, J.R, Brand, S, Smith, V.C, Thompson, S, Smith, A, Davies, K, Mok, N.Y, Torrie, L.S, Collie, I, Hallyburton, I, Norval, S, Simeons, F.R.C, Stojanovski, L, Frearson, J.A, Brenk, R, Wyatt, P.G, Gilbert, I.H, Read, K.D. | | Deposit date: | 2018-05-31 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | A Molecular Hybridization Approach for the Design of Potent, Highly Selective, and Brain-Penetrant N-Myristoyltransferase Inhibitors.
J. Med. Chem., 61, 2018
|
|
6P5M
 
 | | Discovery of a Novel, Highly Potent, and Selective Thieno[3,2-d]pyrimidinone-Based Cdc7 inhibitor with a Quinuclidine Moiety (TAK-931) as an Orally Active Investigational Anti-Tumor Agent | | Descriptor: | 6-(5-methyl-1H-pyrazol-4-yl)-2-[(pyrrolidin-1-yl)methyl]thieno[3,2-d]pyrimidin-4(3H)-one, Rho-associated protein kinase 2 | | Authors: | Hoffman, I.D, Skene, R.J. | | Deposit date: | 2019-05-30 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery of a Novel, Highly Potent, and Selective Thieno[3,2-d]pyrimidinone-Based Cdc7 Inhibitor with a Quinuclidine Moiety (TAK-931) as an Orally Active Investigational Antitumor Agent.
J.Med.Chem., 63, 2020
|
|
6YC4
 
 | | Crystal structure of the steady-state activated state of the light-driven sodium pump KR2 in the pentameric form at room temperature, pH 8.0 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, GLYCEROL, ... | | Authors: | Kovalev, K, Gushchin, I, Gordeliy, V. | | Deposit date: | 2020-03-18 | | Release date: | 2020-04-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular mechanism of light-driven sodium pumping.
Nat Commun, 11, 2020
|
|
6YC1
 
 | | Crystal structure of the H30A mutant of the light-driven sodium pump KR2 in the pentameric form, pH 8.0 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, RETINAL, ... | | Authors: | Kovalev, K, Gushchin, I, Gordeliy, V. | | Deposit date: | 2020-03-18 | | Release date: | 2020-04-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular mechanism of light-driven sodium pumping.
Nat Commun, 11, 2020
|
|
6YFK
 
 | | Virus-like particle of bacteriophage ESE007 | | Descriptor: | CALCIUM ION, coat protein | | Authors: | Rumnieks, J, Kalnins, G, Sisovs, M, Lieknina, I, Tars, K. | | Deposit date: | 2020-03-26 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Three-dimensional structure of 22 uncultured ssRNA bacteriophages: Flexibility of the coat protein fold and variations in particle shapes.
Sci Adv, 6, 2020
|
|
6YFR
 
 | | Virus-like particle of bacteriophage NT-391 | | Descriptor: | coat protein | | Authors: | Rumnieks, J, Kalnins, G, Sisovs, M, Lieknina, I, Tars, K. | | Deposit date: | 2020-03-26 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Three-dimensional structure of 22 uncultured ssRNA bacteriophages: Flexibility of the coat protein fold and variations in particle shapes.
Sci Adv, 6, 2020
|
|
