5HR0
 
 | | Crystal structure of thioredoxin E101G mutant | | Descriptor: | COPPER (II) ION, Thioredoxin | | Authors: | Noguera, M.E, Vazquez, D.S, Howard, E.I, Cousido-Siah, A, Mitschler, A, Podjarny, A, Santos, J. | | Deposit date: | 2016-01-22 | | Release date: | 2017-02-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Structural variability of E. coli thioredoxin captured in the crystal structures of single-point mutants.
Sci Rep, 7, 2017
|
|
6HHK
 
 | |
5J5S
 
 | | Src kinase in complex with a sulfonamide inhibitor | | Descriptor: | N-{4-[8-amino-3-(propan-2-yl)imidazo[1,5-a]pyrazin-1-yl]naphthalen-1-yl}-N'-[3-(trifluoromethyl)phenyl]urea, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Lebedev, I, Feldman, H.C, Georghiou, G, Maly, D.J, Seeliger, M. | | Deposit date: | 2016-04-03 | | Release date: | 2016-06-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.153 Å) | | Cite: | Structural and Functional Analysis of the Allosteric Modulation of IRE1a with ATP-Competitive Kinase Inhibitors
To Be Published
|
|
5LT9
 
 | | Ligand binding domain of Pseudomonas aeruginosa PAO1 amino acid chemoreceptors PctB in complex with L-Arg | | Descriptor: | ARGININE, GLYCEROL, Methyl-accepting chemotaxis protein PctB, ... | | Authors: | Gavira, J.A, Rico-Jimenez, M, Ortega, A, Conejero-Muriel, M, Zhulin, I, Krell, T. | | Deposit date: | 2016-09-06 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | How Bacterial Chemoreceptors Evolve Novel Ligand Specificities
Mbio, 2020
|
|
8UAN
 
 | | The crystal structure of cobalt-bound human ADO C18S C239S variant at 1.99 Angstrom | | Descriptor: | 2-aminoethanethiol dioxygenase, COBALT (II) ION, GLYCEROL | | Authors: | Liu, A, Li, J, Duan, R, Shin, I. | | Deposit date: | 2023-09-21 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Cobalt(II)-Substituted Cysteamine Dioxygenase Oxygenation Proceeds through a Cobalt(III)-Superoxo Complex.
J.Am.Chem.Soc., 146, 2024
|
|
5IGK
 
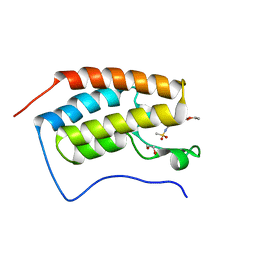 | | Crystal structure of the first bromodomain of human BRD4 in complex with bromosporine (BSP) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, Bromosporine | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, von Delft, F, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-02-28 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Promiscuous targeting of bromodomains by bromosporine identifies BET proteins as master regulators of primary transcription response in leukemia.
Sci Adv, 2, 2016
|
|
5II1
 
 | | Crystal Structure of the fifth bromodomain of human polybromo (PB1) in complex with 1-methylisochromeno[3,4-c]pyrazol-5(3H)-one | | Descriptor: | 1-methyl[2]benzopyrano[3,4-c]pyrazol-5(3H)-one, Protein polybromo-1 | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Myrianthopoulos, V, Mikros, E, von Delft, F, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-03-01 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Discovery and Optimization of a Selective Ligand for the Switch/Sucrose Nonfermenting-Related Bromodomains of Polybromo Protein-1 by the Use of Virtual Screening and Hydration Analysis.
J.Med.Chem., 59, 2016
|
|
8BVC
 
 | |
2L71
 
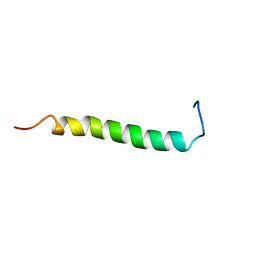 | | NMR solution structure of GIP in Bicellular media | | Descriptor: | Gastric inhibitory polypeptide | | Authors: | Venneti, K.C, Alana, I, O'Harte, F.P.M, Malthouse, P.J.G, Hewage, C.M. | | Deposit date: | 2010-12-01 | | Release date: | 2011-10-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Conformational, receptor interaction and alanine scan studies of glucose-dependent insulinotropic polypeptide
Biochim.Biophys.Acta, 1814, 2011
|
|
2L9Y
 
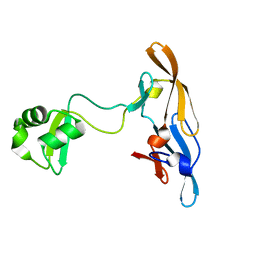 | | Solution structure of the MoCVNH-LysM module from the rice blast fungus Magnaporthe oryzae protein (MGG_03307) | | Descriptor: | CVNH-LysM lectin | | Authors: | Koharudin, L.M.I, Viscomi, A.R, Montanini, B, Kershaw, M.J, Talbot, N.J, Ottonello, S, Gronenborn, A.M. | | Deposit date: | 2011-02-26 | | Release date: | 2011-03-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure-Function Analysis of a CVNH-LysM Lectin Expressed during Plant Infection by the Rice Blast Fungus Magnaporthe oryzae.
Structure, 19, 2011
|
|
6PQA
 
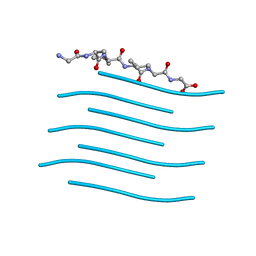 | |
2LSQ
 
 | | Analog of the fragment 197-221 of beta-1 adrenoreceptor | | Descriptor: | ANALOG OF THE FRAGMENT 197-221 OF BETA-1 ADRENORECEPTOR | | Authors: | Bushuev, V.N, Zykov, K.A, Bespalova, Z.D, Bibilashvili, R.S, Efremov, E.E, Golitsyn, S.P, Kaznacheeva, L.I, Kuzhetsova, T.V, Levashov, P.S, Masenko, V.P, Mironova, N.A, Molokoedov, F.S, Rutkevich, P.N, Rvacheva, A.V, Sharf, T.V, Sidorova, M.V, Vlasik, T.N, Yanushevskaya, E.V. | | Deposit date: | 2012-05-04 | | Release date: | 2013-05-08 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Analog of the fragment 197-221 of 1-adrenoreceptor
To be Published
|
|
2L70
 
 | | NMR solution structure of GIP in micellular media | | Descriptor: | Gastric inhibitory polypeptide | | Authors: | Venneti, K.C, Alana, I, O'Harte, F.P.M, Malthouse, P.J.G, Hewage, C.M. | | Deposit date: | 2010-12-01 | | Release date: | 2011-10-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Conformational, receptor interaction and alanine scan studies of glucose-dependent insulinotropic polypeptide
Biochim.Biophys.Acta, 1814, 2011
|
|
2M0K
 
 | |
2M0J
 
 | |
4QSR
 
 | | Structure of the bromodomain of human ATPase family AAA domain-containing protein 2 (ATAD2) with bound MPD | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ATPase family AAA domain-containing protein 2, GLYCEROL, ... | | Authors: | Chaikuad, A, Felletar, I, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-07-06 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based approaches towards identification of fragments for the low-druggability ATAD2 bromodomain
MedChemComm, 5, 2014
|
|
6R45
 
 | | Crystal structure of eukaryotic O-GlcNAcase HAT-like domain | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, O-GlcNAcase | | Authors: | Raimi, O.G, Gorelik, A, Hopkins-Navratilova, I, Aristotelous, T, Ferenbach, A, vanAalten, D. | | Deposit date: | 2019-03-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.784 Å) | | Cite: | Crystal structure of eukaryotic O-GlcNAcase HAT-like domain
To Be Published
|
|
2OZT
 
 | | Crystal structure of O-succinylbenzoate synthase from Thermosynechococcus elongatus BP-1 | | Descriptor: | PHOSPHATE ION, SODIUM ION, Tlr1174 protein | | Authors: | Malashkevich, V.N, Bonanno, J, Toro, R, Sauder, J.M, Schwinn, K.D, Bain, K.T, Adams, J.M, Reyes, C, Rooney, I, Gheyi, T, Wasserman, S.R, Emtage, S, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-02-27 | | Release date: | 2007-03-13 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Loss of quaternary structure is associated with rapid sequence divergence in the OSBS family.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2P6N
 
 | | Human DEAD-box RNA helicase DDX41, helicase domain | | Descriptor: | ATP-dependent RNA helicase DDX41 | | Authors: | Karlberg, T, Ogg, D, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A, Flodin, S, Flores, A, Graslund, S, Hallberg, B.M, Hammarstrom, M, Johansson, I, Kotenyova, T, Lehtio, L, Moche, M, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Stenmark, P, Sundstrom, M, Thorsell, A.G, Van Den Berg, S, Weigelt, J, Holmberg-Schiavone, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-03-19 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Comparative Structural Analysis of Human DEAD-Box RNA Helicases.
Plos One, 5, 2010
|
|
2OX7
 
 | | Crystal structure of protein EF1440 from Enterococcus faecalis | | Descriptor: | Hypothetical protein | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Schwinn, K.D, Thompson, D.A, Bain, K.T, Adams, J.M, Reyes, C, Lau, C, Gilmore, J, Rooney, I, Gheyi, T, Wasserman, S.R, Emtage, S, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-02-19 | | Release date: | 2007-03-06 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.777 Å) | | Cite: | Crystal structure of the hypothetical protein from Enterococcus faecalis
To be Published
|
|
2P84
 
 | | Crystal structure of ORF041 from Bacteriophage 37 | | Descriptor: | ORF041 | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Schwinn, K.D, Thompson, D.A, Bain, K.T, Adams, J.M, Reyes, C, Lau, C, Gilmore, J, Rooney, I, Wasserman, T, Gheyi, S.R, Emtage, S, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-21 | | Release date: | 2007-04-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the hypothetical protein from Staphylococcus phage 37
To be Published
|
|
2PL3
 
 | | Human DEAD-box RNA helicase DDX10, DEAD domain in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ETHANOL, MAGNESIUM ION, ... | | Authors: | Karlberg, T, Lehtio, L, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A, Flodin, S, Flores, A, Graslund, S, Hallberg, B.M, Hammarstrom, M, Johansson, I, Kallas, A, Kotenyova, T, Moche, M, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Stenmark, P, Sundstrom, M, Thorsell, A.G, Van Den Berg, S, Weigelt, J, Holmberg-Schiavone, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-04-19 | | Release date: | 2007-05-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Comparative Structural Analysis of Human DEAD-Box RNA Helicases.
Plos One, 5, 2010
|
|
7BGZ
 
 | | Mutant L39K of recombinant beta-lactoglobulin in complex with endogenous ligand | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactoglobulin, DECANOIC ACID | | Authors: | Loch, J.I, Siuda, M.K, Lewinski, K. | | Deposit date: | 2021-01-09 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Interactions of new lactoglobulin variants with tetracaine: crystallographic studies of ligand binding to lactoglobulin mutants possessing single substitution in the binding pocket.
Acta Biochim.Pol., 68, 2021
|
|
7BF9
 
 | |
7BH0
 
 | |
