1TJR
 
 | | Crystal structure of wild-type BX1 complexed with a sulfate ion | | Descriptor: | BX1, SULFATE ION | | Authors: | Kulik, V, Hartmann, E, Weyand, M, Frey, M, Gierl, A, Niks, D, Dunn, M.F, Schlichting, I. | | Deposit date: | 2004-06-07 | | Release date: | 2005-08-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | On the structural basis of the catalytic mechanism and the regulation of the alpha subunit of tryptophan synthase from Salmonella typhimurium and BX1 from maize, two evolutionarily related enzymes.
J.Mol.Biol., 352, 2005
|
|
3KAR
 
 | | THE MOTOR DOMAIN OF KINESIN-LIKE PROTEIN KAR3, A SACCHAROMYCES CEREVISIAE KINESIN-RELATED PROTEIN | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, KINESIN-LIKE PROTEIN KAR3, MAGNESIUM ION | | Authors: | Gulick, A.M, Song, H, Endow, S, Rayment, I. | | Deposit date: | 1997-11-26 | | Release date: | 1998-04-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray crystal structure of the yeast Kar3 motor domain complexed with Mg.ADP to 2.3 A resolution.
Biochemistry, 37, 1998
|
|
1GH9
 
 | |
1SSF
 
 | | Solution structure of the mouse 53BP1 fragment (residues 1463-1617) | | Descriptor: | Transformation related protein 53 binding protein 1 | | Authors: | Charier, G, Couprie, J, Alpha-Bazin, B, Meyer, V, Quemeneur, E, Guerois, R, Callebaut, I, Gilquin, B, Zinn-Justin, S. | | Deposit date: | 2004-03-24 | | Release date: | 2004-09-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The Tudor Tandem of 53BP1; A New Structural Motif Involved in DNA and RG-Rich Peptide Binding
Structure, 12, 2004
|
|
1SSH
 
 | | Crystal structure of the SH3 domain from a S. cerevisiae hypothetical 40.4 kDa protein in complex with a peptide | | Descriptor: | 12-mer peptide from Cytoskeleton assembly control protein SLA1, Hypothetical 40.4 kDa protein in PES4-HIS2 intergenic region | | Authors: | Kursula, P, Kursula, I, Lehmann, F, Song, Y.-H, Wilmanns, M. | | Deposit date: | 2004-03-24 | | Release date: | 2005-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Yeast SH3 domain structural genomics
To be Published
|
|
1JH8
 
 | | Structural Investigation of the Biosynthesis of Alternative Lower Ligands for Cobamides by Nicotinate Mononucleotide:5,6-Dimethylbenzimidazole Phosphoribosyltransferase (CobT) from Salmonella enterica | | Descriptor: | ADENINE, Nicotinate Mononucleotide:5,6-Dimethylbenzimidazole Phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Cheong, C.-G, Escalante-Semerena, J, Rayment, I. | | Deposit date: | 2001-06-27 | | Release date: | 2001-09-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural investigation of the biosynthesis of alternative lower ligands for cobamides by nicotinate mononucleotide: 5,6-dimethylbenzimidazole phosphoribosyltransferase from Salmonella enterica.
J.Biol.Chem., 276, 2001
|
|
1PBI
 
 | |
1JHP
 
 | | Three-dimensional Structure of CobT in Complex with 5-methoxybenzimidazole | | Descriptor: | 5-METHOXYBENZIMIDAZOLE, Nicotinate Mononucleotide:5,6-Dimethylbenzimidazole Phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Cheong, C.G, Escalante-Semerena, J, Rayment, I. | | Deposit date: | 2001-06-28 | | Release date: | 2001-10-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural investigation of the biosynthesis of alternative lower ligands for cobamides by nicotinate mononucleotide: 5,6-dimethylbenzimidazole phosphoribosyltransferase from Salmonella enterica.
J.Biol.Chem., 276, 2001
|
|
1JHX
 
 | | Three-dimensional Structure of CobT in Complex with Phenol | | Descriptor: | Nicotinate Mononucleotide:5,6-Dimethylbenzimidazole Phosphoribosyltransferase, PHENOL, PHOSPHATE ION | | Authors: | Cheong, C.G, Escalante-Semerena, J, Rayment, I. | | Deposit date: | 2001-06-28 | | Release date: | 2001-10-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural investigation of the biosynthesis of alternative lower ligands for cobamides by nicotinate mononucleotide: 5,6-dimethylbenzimidazole phosphoribosyltransferase from Salmonella enterica.
J.Biol.Chem., 276, 2001
|
|
1JX6
 
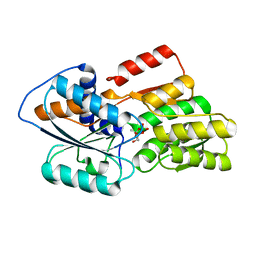 | | CRYSTAL STRUCTURE OF LUXP FROM VIBRIO HARVEYI COMPLEXED WITH AUTOINDUCER-2 | | Descriptor: | 3A-METHYL-5,6-DIHYDRO-FURO[2,3-D][1,3,2]DIOXABOROLE-2,2,6,6A-TETRAOL, CALCIUM ION, LUXP PROTEIN | | Authors: | Chen, X, Schauder, S, Potier, N, Van Dorsselaer, A, Pelczer, I, BassleR, B.L, Hughson, F.M. | | Deposit date: | 2001-09-05 | | Release date: | 2002-02-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural identification of a bacterial quorum-sensing signal containing boron.
Nature, 415, 2002
|
|
1JY9
 
 | | MINIMIZED AVERAGE STRUCTURE OF DP-TT2 | | Descriptor: | DP-TT2 | | Authors: | Stanger, H.E, Syud, F.A, Espinosa, J.F, Giriat, I, Muir, T, Gellman, S.H. | | Deposit date: | 2001-09-11 | | Release date: | 2001-09-19 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Length-dependent stability and strand length limits in antiparallel beta -sheet secondary structure.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1JYD
 
 | | Crystal Structure of Recombinant Human Serum Retinol-Binding Protein at 1.7 A Resolution | | Descriptor: | GLYCEROL, PLASMA RETINOL-BINDING PROTEIN | | Authors: | Greene, L.H, Chrysina, E.D, Irons, L.I, Papageorgiou, A.C, Acharya, K.R, Brew, K. | | Deposit date: | 2001-09-12 | | Release date: | 2003-07-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Role of conserved residues in structure and stability: Tryptophans of human serum
retinol-binding protein, a model for the lipocalin superfamily
Protein Sci., 10, 2001
|
|
2UYQ
 
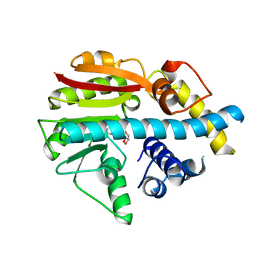 | | Crystal structure of ML2640c from Mycobacterium leprae in complex with S-adenosylmethionine | | Descriptor: | HYPOTHETICAL PROTEIN ML2640, S-ADENOSYLMETHIONINE | | Authors: | Grana, M, Buschiazzo, A, Wehenkel, A, Haouz, A, Miras, I, Shepard, W, Alzari, P.M. | | Deposit date: | 2007-04-11 | | Release date: | 2007-08-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Crystal Structure of M. Leprae Ml2640C Defines a Large Family of Putative S-Adenosylmethionine- Dependent Methyltransferases in Mycobacteria.
Protein Sci., 16, 2007
|
|
1JJF
 
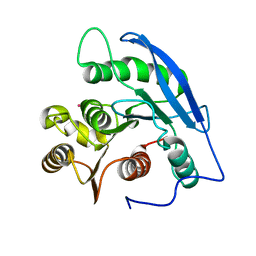 | | STRUCTURAL BASIS FOR THE SUBSTRATE SPECIFICITY OF THE FERULOYL ESTERASE DOMAIN OF THE CELLULOSOMAL XYLANASE Z OF CLOSTRIDIUM THERMOCELLUM | | Descriptor: | ENDO-1,4-BETA-XYLANASE Z, PLATINUM (II) ION | | Authors: | Schubot, F.D, Kataeva, I.A, Blum, D.L, Shah, A.K, Ljungdahl, L.G, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2001-07-05 | | Release date: | 2001-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for the substrate specificity of the feruloyl esterase domain of the cellulosomal xylanase Z from Clostridium thermocellum.
Biochemistry, 40, 2001
|
|
2VBD
 
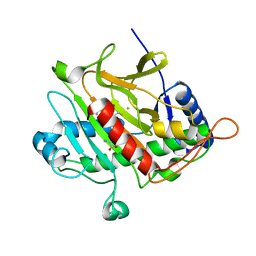 | | Isopenicillin N synthase with substrate analogue L,L,L-ACOMP (unexposed) | | Descriptor: | FE (II) ION, ISOPENICILLIN N SYNTHETASE, N^6^-[(1R)-2-[(1R)-1-carboxy-2-(methylsulfanyl)ethoxy]-2-oxo-1-(sulfanylmethyl)ethyl]-6-oxo-L-lysine | | Authors: | Ge, W, Clifton, I.J, Adlington, R.M, Baldwin, J.E, Rutledge, P.J. | | Deposit date: | 2007-09-10 | | Release date: | 2008-09-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of an Lll-Configured Depsipeptide Substrate Analogue Bound to Isopenicillin N Synthase.
Org.Biomol.Chem., 8, 2010
|
|
3U2M
 
 | | Crystal structure of human ALR mutant C142/145S | | Descriptor: | FAD-linked sulfhydryl oxidase ALR, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Banci, L, Bertini, I, Calderone, V, Cefaro, C, Ciofi-Baffoni, S, Gallo, A. | | Deposit date: | 2011-10-04 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An electron-transfer path through an extended disulfide relay system: the case of the redox protein ALR.
J.Am.Chem.Soc., 134, 2012
|
|
2UZ9
 
 | | Human guanine deaminase (guaD) in complex with zinc and its product Xanthine. | | Descriptor: | GUANINE DEAMINASE, XANTHINE, ZINC ION | | Authors: | Moche, M, Welin, M, Arrowsmith, C, Berglund, H, Busam, R, Collins, R, Dahlgren, L.G, Edwards, A, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Holmberg-Schiavone, L, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Lehtio, L, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Stenmark, P, Sundstrom, M, Thorsell, A.G, van den berg, S, Weigelt, J, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-04-26 | | Release date: | 2007-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Human Guanine Deaminase (Guad) in Complex with Zinc and its Product Xhantine
To be Published
|
|
2UYO
 
 | | Crystal structure of ML2640c from Mycobacterium leprae in an hexagonal crystal form | | Descriptor: | HYPOTHETICAL PROTEIN ML2640 | | Authors: | Grana, M, Buschiazzo, A, Wehenkel, A, Haouz, A, Miras, I, Shepard, W, Alzari, P.M. | | Deposit date: | 2007-04-11 | | Release date: | 2007-08-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of M. Leprae Ml2640C Defines a Large Family of Putative S-Adenosylmethionine- Dependent Methyltransferases in Mycobacteria.
Protein Sci., 16, 2007
|
|
2V58
 
 | | CRYSTAL STRUCTURE OF BIOTIN CARBOXYLASE FROM E.COLI IN COMPLEX WITH POTENT INHIBITOR 1 | | Descriptor: | 6-(2,6-dibromophenyl)pyrido[2,3-d]pyrimidine-2,7-diamine, BIOTIN CARBOXYLASE, CHLORIDE ION | | Authors: | Mochalkin, I, Miller, J.R. | | Deposit date: | 2008-10-02 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Class of Selective Antibacterials Derived from a Protein Kinase Inhibitor Pharmacophore.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2VBB
 
 | | Isopenicillin N synthase with substrate analogue ACOMP (35minutes oxygen exposure) | | Descriptor: | FE (II) ION, GLYCEROL, ISOPENICILLIN N SYNTHETASE, ... | | Authors: | Ge, W, Clifton, I.J, Adlington, R.M, Baldwin, J.E, Rutledge, P.J. | | Deposit date: | 2007-09-07 | | Release date: | 2008-07-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Isopenicillin N Synthase Mediates Thiolate Oxidation to Sulfenate in a Depsipeptide Substrate Analogue: Implications for Oxygen Binding and a Link to Nitrile Hydratase?
J.Am.Chem.Soc., 130, 2008
|
|
2VFB
 
 | | The structure of Mycobacterium marinum arylamine N-acetyltransferase | | Descriptor: | ARYLAMINE N-ACETYLTRANSFERASE | | Authors: | Fullam, E, Westwood, I.M, Anderton, M.C, Lowe, E.D, Sim, E, Noble, M.E.M. | | Deposit date: | 2007-11-02 | | Release date: | 2007-12-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Divergence of Cofactor Recognition Across Evolution: Coenzyme a Binding in a Prokaryotic Arylamine N-Acetyltransferase.
J.Mol.Biol., 375, 2008
|
|
1K2I
 
 | | Crystal Structure of Gamma-Chymotrypsin in Complex with 7-Hydroxycoumarin | | Descriptor: | 2,4-DIHYDROXY-TRANS CINNAMIC ACID, CHYMOTRYPSINOGEN A, SULFATE ION | | Authors: | Ghani, U, Ng, K.K.S, Atta-ur-Rahman, Choudhary, M.I, Ullah, N, James, M.N.G. | | Deposit date: | 2001-09-27 | | Release date: | 2001-12-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of gamma-chymotrypsin in complex with 7-hydroxycoumarin.
J.Mol.Biol., 314, 2001
|
|
1JGV
 
 | | STRUCTURAL BASIS FOR DISFAVORED ELIMINATION REACTION IN CATALYTIC ANTIBODY 1D4 | | Descriptor: | Antibody Heavy Chain, Antibody Light Chain | | Authors: | Larsen, N.A, Heine, A, Crane, L, Cravatt, B.F, Lerner, R.A, Wilson, I.A. | | Deposit date: | 2001-06-26 | | Release date: | 2001-12-05 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for a disfavored elimination reaction in catalytic antibody 1D4.
J.Mol.Biol., 314, 2001
|
|
1JH6
 
 | | Semi-reduced Cyclic Nucleotide Phosphodiesterase from Arabidopsis thaliana | | Descriptor: | SULFATE ION, cyclic phosphodiesterase | | Authors: | Hofmann, A, Grella, M, Botos, I, Filipowicz, W, Wlodawer, A. | | Deposit date: | 2001-06-27 | | Release date: | 2002-02-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the semireduced and inhibitor-bound forms of cyclic nucleotide phosphodiesterase from Arabidopsis thaliana.
J.Biol.Chem., 277, 2002
|
|
2VKT
 
 | | HUMAN CTP SYNTHETASE 2 - GLUTAMINASE DOMAIN | | Descriptor: | CTP SYNTHASE 2 | | Authors: | Welin, M, Tresaugues, L, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Herman, M.D, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Lehtio, L, Moche, M, Nilsson, M.E, Nyman, T, Persson, C, Sagemark, J, Svensson, L, Thorsell, A.G, van den Berg, S, Weigelt, J, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-12-28 | | Release date: | 2008-01-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Human Ctp Synthetase 2 - Glutaminase Domain
To be Published
|
|
