1N83
 
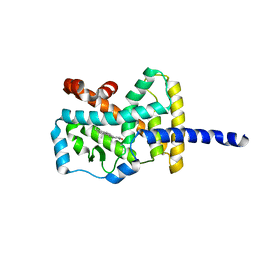 | | Crystal Structure of the complex between the Orphan Nuclear Hormone Receptor ROR(alpha)-LBD and Cholesterol | | Descriptor: | CHOLESTEROL, Nuclear receptor ROR-alpha | | Authors: | Kallen, J.A, Schlaeppi, J.M, Bitsch, F, Geisse, S, Geiser, M, Delhon, I, Fournier, B. | | Deposit date: | 2002-11-19 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | X-ray Structure of hROR(alpha) LBD at 1.63A: Structural and Functional data that Cholesterol or a Cholesterol derivative is the natural ligand of ROR(alpha)
Structure, 10, 2002
|
|
1N8M
 
 | | Solution structure of Pi4, a four disulfide bridged scorpion toxin active on potassium channels | | Descriptor: | Potassium channel blocking toxin 4 | | Authors: | Guijarro, J.I, M'Barek, S, Olamendi-Portugal, T, Gomez-Lagunas, F, Garnier, D, Rochat, H, Possani, L.D, Sabatier, J.M, Delepierre, M. | | Deposit date: | 2002-11-21 | | Release date: | 2003-09-02 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Pi4, a short four-disulfide-bridged scorpion toxin specific of potassium channels.
Protein Sci., 12, 2003
|
|
1JGV
 
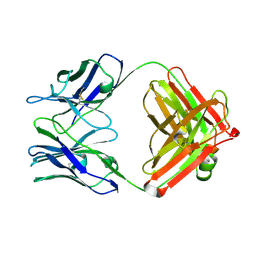 | | STRUCTURAL BASIS FOR DISFAVORED ELIMINATION REACTION IN CATALYTIC ANTIBODY 1D4 | | Descriptor: | Antibody Heavy Chain, Antibody Light Chain | | Authors: | Larsen, N.A, Heine, A, Crane, L, Cravatt, B.F, Lerner, R.A, Wilson, I.A. | | Deposit date: | 2001-06-26 | | Release date: | 2001-12-05 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for a disfavored elimination reaction in catalytic antibody 1D4.
J.Mol.Biol., 314, 2001
|
|
1JH6
 
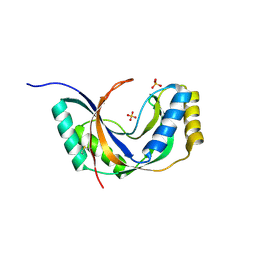 | | Semi-reduced Cyclic Nucleotide Phosphodiesterase from Arabidopsis thaliana | | Descriptor: | SULFATE ION, cyclic phosphodiesterase | | Authors: | Hofmann, A, Grella, M, Botos, I, Filipowicz, W, Wlodawer, A. | | Deposit date: | 2001-06-27 | | Release date: | 2002-02-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the semireduced and inhibitor-bound forms of cyclic nucleotide phosphodiesterase from Arabidopsis thaliana.
J.Biol.Chem., 277, 2002
|
|
1N9O
 
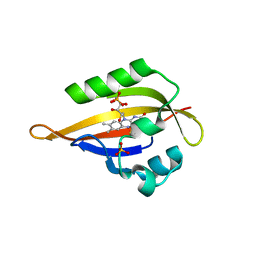 | | Crystal structure of the Phot-LOV1 domain from Chlamydomonas reinhardtii in illuminated state. Composite data set. | | Descriptor: | FLAVIN MONONUCLEOTIDE, SULFATE ION, putative blue light receptor | | Authors: | Fedorov, R, Schlichting, I, Hartmann, E, Domratcheva, T, Fuhrmann, M, Hegemann, P. | | Deposit date: | 2002-11-25 | | Release date: | 2003-06-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures and molecular mechanism of a light-induced signaling switch: The Phot-LOV1 domain from Chlamydomonas reinhardtii.
Biophys.J., 84, 2003
|
|
1JTH
 
 | | Crystal structure and biophysical properties of a complex between the N-terminal region of SNAP25 and the SNARE region of syntaxin 1a | | Descriptor: | SNAP25, syntaxin 1a | | Authors: | Misura, K.M.S, Gonzalez Jr, L.C, May, A.P, Scheller, R.H, Weis, W.I. | | Deposit date: | 2001-08-21 | | Release date: | 2001-11-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and biophysical properties of a complex between the N-terminal SNARE region of SNAP25 and syntaxin 1a.
J.Biol.Chem., 276, 2001
|
|
7L65
 
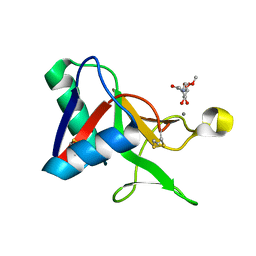 | |
7L68
 
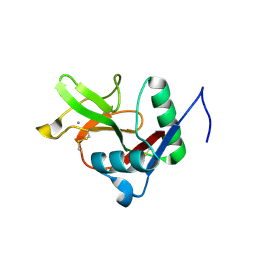 | |
1JT2
 
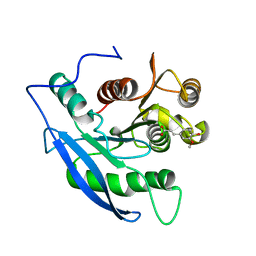 | | STRUCTURAL BASIS FOR THE SUBSTRATE SPECIFICITY OF THE FERUL DOMAIN OF THE CELLULOSOMAL XYLANASE Z FROM C. THERMOCELLUM | | Descriptor: | 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, PROTEIN (ENDO-1,4-BETA-XYLANASE Z) | | Authors: | Schubot, F.D, Kataeva, I.A, Blum, D.L, Shah, A.K, Ljungdahl, L.G, Rose, J.P, Wang, B.-C. | | Deposit date: | 2001-08-20 | | Release date: | 2002-03-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the substrate specificity of the feruloyl esterase domain of the cellulosomal xylanase Z from Clostridium thermocellum.
Biochemistry, 40, 2001
|
|
1JT3
 
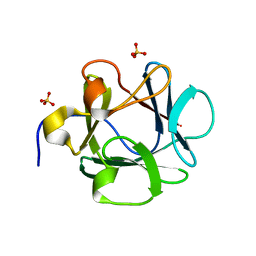 | | Human Acidic Fibroblast Growth Factor. 141 Amino Acid Form with Amino Histidine Tag AND LEU 73 REPLACED BY VAL (L73V) | | Descriptor: | SULFATE ION, acidic fibroblast growth factor | | Authors: | Brych, S.R, Blaber, S.I, Logan, T.M, Blaber, M. | | Deposit date: | 2001-08-20 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and stability effects of mutations designed to increase the primary sequence symmetry within the core
region of a beta-trefoil.
Protein Sci., 10, 2001
|
|
1K1G
 
 | | STRUCTURAL BASIS FOR RECOGNITION OF THE INTRON BRANCH SITE RNA BY SPLICING FACTOR 1 | | Descriptor: | 5'-R(*UP*AP*UP*AP*CP*UP*AP*AP*CP*AP*A)-3', SF1-Bo isoform | | Authors: | Liu, Z, Luyten, I, Bottomley, M.J, Messias, A.C, Houngninou-Molango, S, Sprangers, R, Zanier, K, Kramer, A, Sattler, M. | | Deposit date: | 2001-09-25 | | Release date: | 2001-11-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis for recognition of the intron branch site RNA by splicing factor 1.
Science, 294, 2001
|
|
1XMJ
 
 | | Crystal structure of human deltaF508 human NBD1 domain with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION | | Authors: | Lewis, H.A, Zhao, X, Wang, C, Sauder, J.M, Rooney, I, Noland, B.W, Lorimer, D, Kearins, M.C, Conners, K, Condon, B, Maloney, P.C, Guggino, W.B, Hunt, J.F, Emtage, S, Structural GenomiX | | Deposit date: | 2004-10-02 | | Release date: | 2004-11-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Impact of the delta-F508 Mutation in First Nucleotide-binding Domain of Human Cystic Fibrosis Transmembrane Conductance Regulator on Domain Folding and Structure
J.Biol.Chem., 280, 2005
|
|
1XBY
 
 | | Structure of 3-keto-L-gulonate 6-phosphate decarboxylase E112D/T169A mutant with bound D-ribulose 5-phosphate | | Descriptor: | 3-keto-L-gulonate 6-phosphate decarboxylase, MAGNESIUM ION, RIBULOSE-5-PHOSPHATE | | Authors: | Wise, E.L, Yew, W.S, Akana, J, Gerlt, J.A, Rayment, I. | | Deposit date: | 2004-08-31 | | Release date: | 2005-04-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Evolution of enzymatic activities in the orotidine 5'-monophosphate decarboxylase suprafamily: structural basis for catalytic promiscuity in wild-type and designed mutants of 3-keto-L-gulonate 6-phosphate decarboxylase
Biochemistry, 44, 2005
|
|
1OHB
 
 | | Acetylglutamate kinase from Escherichia coli complexed with ADP and sulphate | | Descriptor: | ACETATE ION, ACETYLGLUTAMATE KINASE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Gil-Ortiz, F, Ramon-Maiques, S, Fita, I, Rubio, V. | | Deposit date: | 2003-05-23 | | Release date: | 2003-07-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Course of Phosphorus in the Reaction of N-Acetyl-L-Glutamate Kinase, Determined from the Structures of Crystalline Complexes, Including a Complex with an Alf(4)(-) Transition State Mimic
J.Mol.Biol., 331, 2003
|
|
1XBZ
 
 | | Crystal structure of 3-keto-L-gulonate 6-phosphate decarboxylase E112D/R139V/T169A mutant with bound L-xylulose 5-phosphate | | Descriptor: | 3-Keto-L-Gulonate 6-Phosphate Decarboxylase, L-XYLULOSE 5-PHOSPHATE, MAGNESIUM ION | | Authors: | Wise, E.L, Yew, W.S, Akana, J, Gerlt, J.A, Rayment, I. | | Deposit date: | 2004-08-31 | | Release date: | 2005-04-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evolution of enzymatic activities in the orotidine 5'-monophosphate decarboxylase suprafamily: structural basis for catalytic promiscuity in wild-type and designed mutants of 3-keto-L-gulonate 6-phosphate decarboxylase
Biochemistry, 44, 2005
|
|
1JJB
 
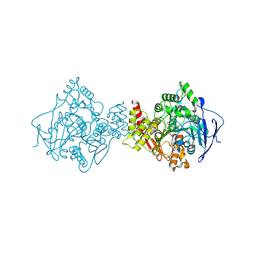 | | A neutral molecule in cation-binding site: Specific binding of PEG-SH to Acetylcholinesterase from Torpedo californica | | Descriptor: | 1-DEOXY-1-THIO-HEPTAETHYLENE GLYCOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE | | Authors: | Koellner, G, Steiner, T, Millard, C.B, Silman, I, Sussman, J.L. | | Deposit date: | 2001-07-04 | | Release date: | 2002-07-17 | | Last modified: | 2021-06-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A neutral molecule in a cation-binding site: specific binding of a PEG-SH to acetylcholinesterase from Torpedo californica.
J.Mol.Biol., 320, 2002
|
|
3KYJ
 
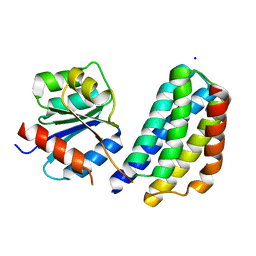 | | Crystal structure of the P1 domain of CheA3 in complex with CheY6 from R. sphaeroides | | Descriptor: | CheY6 protein, Putative histidine protein kinase, SODIUM ION | | Authors: | Bell, C.H, Porter, S.L, Armitage, J.P, Stuart, D.I. | | Deposit date: | 2009-12-06 | | Release date: | 2010-02-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Using structural information to change the phosphotransfer specificity of a two-component chemotaxis signalling complex
Plos Biol., 8, 2010
|
|
1XDC
 
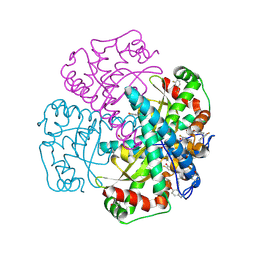 | | Hydrogen Bonding in Human Manganese Superoxide Dismutase containing 3-Fluorotyrosine | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase [Mn], mitochondrial | | Authors: | Ayala, I, Perry, J.J, Szczepanski, J, Cabelli, D.E, Tainer, J.A, Vala, M.T, Nick, H.S, Silverman, D.N. | | Deposit date: | 2004-09-05 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Hydrogen bonding in human manganese superoxide dismutase containing 3-fluorotyrosine
Biophys.J., 89, 2005
|
|
1XGW
 
 | |
7KRK
 
 | |
1OH9
 
 | | Acetylglutamate kinase from Escherichia coli complexed with MgADP, N-acetyl-L-glutamate and the transition-state mimic AlF4- | | Descriptor: | ACETYLGLUTAMATE KINASE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Gil-Ortiz, F, Ramon-Maiques, S, Fita, I, Rubio, V. | | Deposit date: | 2003-05-23 | | Release date: | 2003-07-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | The Course of Phosphorus in the Reaction of N-Acetyl-L-Glutamate Kinase, Determined from the Structures of Crystalline Complexes, Including a Complex with an Alf(4)(-) Transition State Mimic
J.Mol.Biol., 331, 2003
|
|
1K2B
 
 | | Combining Mutations in HIV-1 Protease to Understand Mechanisms of Resistance | | Descriptor: | N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide, PROTEASE RETROPEPSIN | | Authors: | Mahalingam, B, Boross, P, Wang, Y.-F, Louis, J.M, Fischer, C, Tozser, J, W Harrison, R, Weber, I.T. | | Deposit date: | 2001-09-26 | | Release date: | 2002-07-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Combining mutations in HIV-1 protease to understand mechanisms of resistance.
Proteins, 48, 2002
|
|
1XGV
 
 | | Isocitrate Dehydrogenase from the hyperthermophile Aeropyrum pernix | | Descriptor: | Isocitrate dehydrogenase | | Authors: | Karlstrom, M, Stokke, R, Steen, I.H, Birkeland, N.-K, Ladenstein, R. | | Deposit date: | 2004-09-17 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Isocitrate dehydrogenase from the hyperthermophile Aeropyrum pernix: X-ray structure analysis of a ternary enzyme-substrate complex and thermal stability
J.Mol.Biol., 345, 2005
|
|
1JN5
 
 | | Structural basis for the recognition of a nucleoporin FG-repeat by the NTF2-like domain of TAP-p15 mRNA export factor | | Descriptor: | FG-repeat, TAP, p15 | | Authors: | Fribourg, S, Braun, I.C, Izaurralde, E, Conti, E. | | Deposit date: | 2001-07-23 | | Release date: | 2001-10-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the recognition of a nucleoporin FG repeat by the NTF2-like domain of the TAP/p15 mRNA nuclear export factor.
Mol.Cell, 8, 2001
|
|
7L61
 
 | |
