7BET
 
 | | Structure of Ribonucleotide reductase R2 from Escherichia coli collected by femtosecond serial crystallography on a COC membrane | | Descriptor: | FE (III) ION, Ribonucleoside-diphosphate reductase 1 subunit beta | | Authors: | Aurelius, O, John, J, Martiel, I, Marsh, M, Vera, L, Huang, C.Y, Olieric, V, Leonarski, P, Nass, K, Padeste, C, Karpik, A, Hogbom, M, Wang, M, Pedrini, B. | | Deposit date: | 2020-12-24 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Commissioning results from the SwissMX instrument for fixed target macromolecular crystallography at SwissFEL
To Be Published
|
|
1BIT
 
 | |
1U8A
 
 | | Crystal Structure of Mycobacterium Tuberculosis Shikimate Kinase in Complex with Shikimate and ADP at 2.15 Angstrom Resolution | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Dhaliwal, B, Nichols, C.E, Ren, J, Lockyer, M, Charles, I, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2004-08-05 | | Release date: | 2004-10-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystallographic studies of shikimate binding and induced conformational changes in Mycobacterium tuberculosis shikimate kinase.
Febs Lett., 574, 2004
|
|
4HJU
 
 | |
1BIH
 
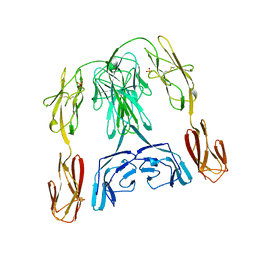 | | CRYSTAL STRUCTURE OF THE INSECT IMMUNE PROTEIN HEMOLIN: A NEW DOMAIN ARRANGEMENT WITH IMPLICATIONS FOR HOMOPHILIC ADHESION | | Descriptor: | HEMOLIN, PHOSPHATE ION | | Authors: | Su, X.-D, Gastinel, L.N, Vaughn, D.E, Faye, I, Poon, P, Bjorkman, P.J. | | Deposit date: | 1998-06-17 | | Release date: | 1998-10-14 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of hemolin: a horseshoe shape with implications for homophilic adhesion.
Science, 281, 1998
|
|
1TR4
 
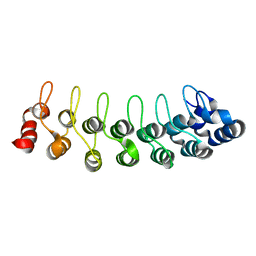 | | Solution structure of human oncogenic protein gankyrin | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 10 | | Authors: | Yuan, C, Li, J, Mahajan, A, Poi, M.J, Byeon, I.J, Tsai, M.D. | | Deposit date: | 2004-06-19 | | Release date: | 2004-11-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the human oncogenic protein gankyrin containing seven ankyrin repeats and analysis of its structure--function relationship.
Biochemistry, 43, 2004
|
|
1BE0
 
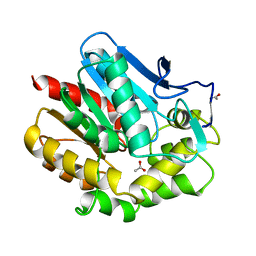 | | HALOALKANE DEHALOGENASE AT PH 5.0 CONTAINING ACETIC ACID | | Descriptor: | ACETATE ION, ACETIC ACID, HALOALKANE DEHALOGENASE | | Authors: | Ridder, I.S, Vos, G.J, Rozeboom, H.J, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 1998-05-18 | | Release date: | 1998-11-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Kinetic analysis and X-ray structure of haloalkane dehalogenase with a modified halide-binding site.
Biochemistry, 37, 1998
|
|
1TTX
 
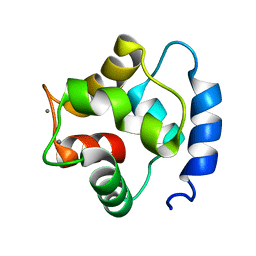 | | Solution Structure of human beta parvalbumin (oncomodulin) refined with a paramagnetism based strategy | | Descriptor: | CALCIUM ION, Oncomodulin | | Authors: | Babini, E, Bertini, I, Capozzi, F, Del Bianco, C, Hollender, D, Kiss, T, Luchinat, C, Quattrone, A, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-06-23 | | Release date: | 2005-01-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Human beta-Parvalbumin and Structural Comparison with Its Paralog alpha-Parvalbumin and with Their Rat Orthologs(,)
Biochemistry, 43, 2004
|
|
3KYJ
 
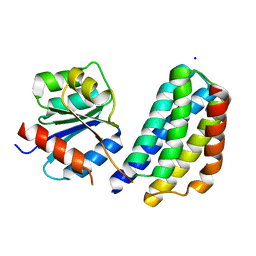 | | Crystal structure of the P1 domain of CheA3 in complex with CheY6 from R. sphaeroides | | Descriptor: | CheY6 protein, Putative histidine protein kinase, SODIUM ION | | Authors: | Bell, C.H, Porter, S.L, Armitage, J.P, Stuart, D.I. | | Deposit date: | 2009-12-06 | | Release date: | 2010-02-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Using structural information to change the phosphotransfer specificity of a two-component chemotaxis signalling complex
Plos Biol., 8, 2010
|
|
1BHA
 
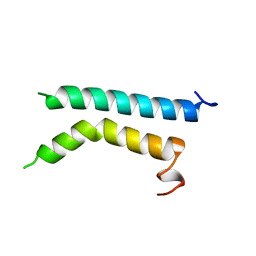 | | THREE-DIMENSIONAL STRUCTURE OF (1-71) BACTERIOOPSIN SOLUBILIZED IN METHANOL-CHLOROFORM AND SDS MICELLES DETERMINED BY 15N-1H HETERONUCLEAR NMR SPECTROSCOPY | | Descriptor: | BACTERIORHODOPSIN | | Authors: | Pervushin, K.V, Orekhov, V.Y, Popov, A.I, Musina, L.Y, Arseniev, A.S. | | Deposit date: | 1993-10-11 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of (1-71)bacterioopsin solubilized in methanol/chloroform and SDS micelles determined by 15N-1H heteronuclear NMR spectroscopy.
Eur.J.Biochem., 219, 1994
|
|
1BGF
 
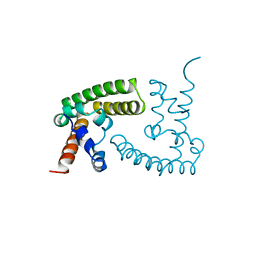 | |
1BHB
 
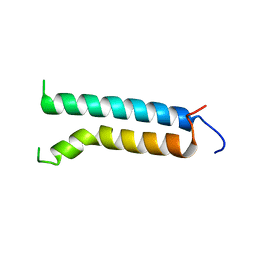 | | Three-dimensional structure of (1-71) bacterioopsin solubilized in methanol-chloroform and SDS micelles determined by 15N-1H heteronuclear NMR spectroscopy | | Descriptor: | BACTERIORHODOPSIN | | Authors: | Orekhov, V.Y, Pervushin, K.V, Popov, A.I, Musina, L.Y, Arseniev, A.S. | | Deposit date: | 1993-10-11 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of (1-71)bacterioopsin solubilized in methanol/chloroform and SDS micelles determined by 15N-1H heteronuclear NMR spectroscopy.
Eur.J.Biochem., 219, 1994
|
|
3KCZ
 
 | | Human poly(ADP-ribose) polymerase 2, catalytic fragment in complex with an inhibitor 3-aminobenzamide | | Descriptor: | 3-aminobenzamide, GLYCEROL, Poly [ADP-ribose] polymerase 2 | | Authors: | Karlberg, T, Schutz, P, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kallas, A, Kotenyova, T, Kotzsch, A, Kraulis, P, Nielsen, T.K, Moche, M, Nordlund, P, Nyman, T, Persson, C, Roos, A.K, Siponen, M.I, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Wisniewska, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-10-22 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the catalytic domain of human PARP2 in complex with PARP inhibitor ABT-888.
Biochemistry, 49, 2010
|
|
1U4Q
 
 | | Crystal Structure of Repeats 15, 16 and 17 of Chicken Brain Alpha Spectrin | | Descriptor: | Spectrin alpha chain, brain | | Authors: | Kusunoki, H, Minasov, G, MacDonald, R.I, Mondragon, A. | | Deposit date: | 2004-07-26 | | Release date: | 2004-10-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Independent Movement, Dimerization and Stability of Tandem Repeats of Chicken Brain alpha-Spectrin
J.Mol.Biol., 344, 2004
|
|
1TZV
 
 | | T. maritima NusB, P3121, Form 1 | | Descriptor: | N utilization substance protein B homolog | | Authors: | Bonin, I, Robelek, R, Benecke, H, Urlaub, H, Bacher, A, Richter, G, Wahl, M.C. | | Deposit date: | 2004-07-12 | | Release date: | 2004-08-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structures of the antitermination factor NusB from Thermotoga maritima and implications for RNA binding
Biochem.J., 383, 2004
|
|
1UA5
 
 | |
4HJT
 
 | |
3KGU
 
 | |
1BFY
 
 | | SOLUTION STRUCTURE OF REDUCED CLOSTRIDIUM PASTEURIANUM RUBREDOXIN, NMR, 20 STRUCTURES | | Descriptor: | FE (III) ION, RUBREDOXIN | | Authors: | Bertini, I, Kurtz Junior, D.M, Eidsness, M.K, Liu, G, Luchinat, C, Rosato, A, Scott, R.A. | | Deposit date: | 1998-05-23 | | Release date: | 1999-05-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Reduced Clostridium Pasteurianum Rubredoxin
J.Biol.Inorg.Chem., 3, 1998
|
|
7C51
 
 | |
1TQH
 
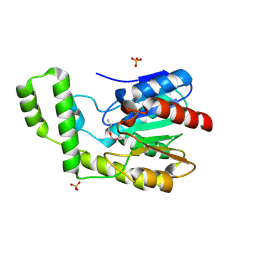 | | Covalent Reaction intermediate Revealed in Crystal Structure of the Geobacillus stearothermophilus Carboxylesterase Est30 | | Descriptor: | Carboxylesterase precursor, PROPYL ACETATE, SULFATE ION | | Authors: | Liu, P, Wang, Y.F, Ewis, H.E, Abdelal, A.T, Lu, C.D, Harrison, R.W, Weber, I.T. | | Deposit date: | 2004-06-17 | | Release date: | 2004-09-28 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Covalent reaction intermediate revealed in crystal structure of the Geobacillus stearothermophilus carboxylesterase Est30.
J.Mol.Biol., 342, 2004
|
|
3KHE
 
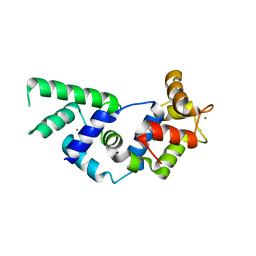 | | Crystal structure of the calcium-loaded calmodulin-like domain of the CDPK, 541.m00134 from toxoplasma gondii | | Descriptor: | CALCIUM ION, Calmodulin-like domain protein kinase isoform 3, GLYCEROL, ... | | Authors: | Wernimont, A.K, Hutchinson, A, Artz, J.D, Mackenzie, F, Cossar, D, Kozieradzki, I, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Hui, R, Qiu, W, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-10-30 | | Release date: | 2010-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of parasitic CDPK domains point to a common mechanism of activation.
Proteins, 79, 2011
|
|
1TQR
 
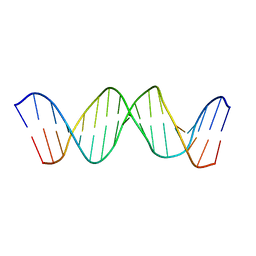 | | NMR Structure of DNA 17-mer GGAAAATCTCTAGCAGT corresponding to the extremity of the U5 LTR of the HIV-1 genome | | Descriptor: | DNA HIV-1 U5 LTR extremity | | Authors: | Renisio, J.G, Cosquer, S, Cherrak, I, El Antri, S, Mauffret, O, Fermandjian, S. | | Deposit date: | 2004-06-18 | | Release date: | 2005-04-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Pre-organized structure of viral DNA at the binding-processing site of HIV-1 integrase
NUCLEIC ACIDS RES., 33, 2005
|
|
1TR6
 
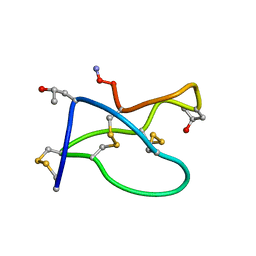 | | NMR solution structure of omega-conotoxin [K10]GVIA, a cyclic cysteine knot peptide | | Descriptor: | Omega-conotoxin GVIA | | Authors: | Mould, J, Yasuda, T, Schroeder, C.I, Beedle, A.M, Doering, C.J, Zamponi, G.W, Adams, D.J, Lewis, R.J. | | Deposit date: | 2004-06-21 | | Release date: | 2004-07-13 | | Last modified: | 2011-10-05 | | Method: | SOLUTION NMR | | Cite: | The alpha2delta auxiliary subunit reduces affinity of omega-conotoxins for recombinant N-type (Cav2.2) calcium channels
J.Biol.Chem., 279, 2004
|
|
1TYO
 
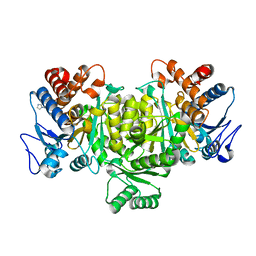 | | Isocitrate Dehydrogenase from the hyperthermophile Aeropyrum pernix in complex with etheno-NADP | | Descriptor: | ETHENO-NADP, isocitrate dehydrogenase | | Authors: | Karlstrom, M, Stokke, R, Steen, I.H, Birkeland, N, Ladenstein, R. | | Deposit date: | 2004-07-08 | | Release date: | 2005-07-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Isocitrate dehydrogenase from the hyperthermophile Aeropyrum pernix: X-ray structure analysis of a ternary enzyme-substrate complex and thermal stability
J.Mol.Biol., 345, 2005
|
|
