7ZRH
 
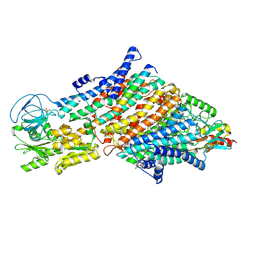 | | Cryo-EM structure of the KdpFABC complex in a nucleotide-free E1 conformation loaded with K+ | | Descriptor: | CARDIOLIPIN, POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, ... | | Authors: | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Stansfeld, P.J, Haenelt, I, Paulino, C. | | Deposit date: | 2022-05-04 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
8YU6
 
 | | The structure of thiocyanate dehydrogenase mutant with the H447Q substitution from Pelomicrobium methylotrophicum (pmTcDH H447Q), activated by crystal soaking with 1mM CuCl2 and 1 mM sodium ascorbate | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Varfolomeeva, L.A, Shipkov, N.S, Dergousova, N.I, Boyko, K.M, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2024-03-26 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The structure of thiocyanate dehydrogenase mutant with the H447Q substitution from Pelomicrobium methylotrophicum (pmTcDH H447Q), activated by crystal soaking with 1mM CuCl2 and 1 mM sodium ascorbate
To Be Published
|
|
7ZRJ
 
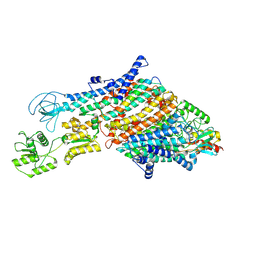 | | Cryo-EM structure of the KdpFABC complex in a nucleotide-free E1 conformation loaded with K+ | | Descriptor: | CARDIOLIPIN, POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, ... | | Authors: | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Stansfeld, P.J, Haenelt, I, Paulino, C. | | Deposit date: | 2022-05-04 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
7ZRG
 
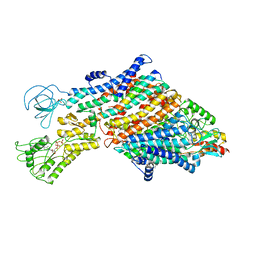 | | Cryo-EM map of the WT KdpFABC complex in the E1_ATPearly conformation, under turnover conditions | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CARDIOLIPIN, POTASSIUM ION, ... | | Authors: | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Rheinberger, J, Wunnicke, D, Dubach, V.R.A, Stansfeld, P.J, Haenelt, I, Paulino, C. | | Deposit date: | 2022-05-04 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
7ZRI
 
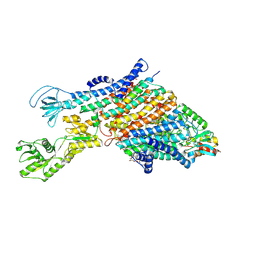 | | Cryo-EM structure of the KdpFABC complex in a nucleotide-free E1 conformation loaded with K+ | | Descriptor: | CARDIOLIPIN, POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, ... | | Authors: | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Stansfeld, P.J, Haenelt, I, Paulino, C. | | Deposit date: | 2022-05-04 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
8A22
 
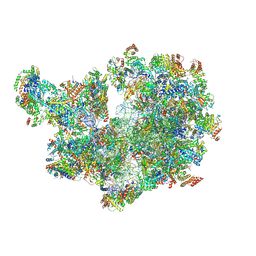 | | Structure of the mitochondrial ribosome from Polytomella magna | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Tobiasson, V, Berzina, I, Amunts, A. | | Deposit date: | 2022-06-02 | | Release date: | 2022-11-16 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structure of a mitochondrial ribosome with fragmented rRNA in complex with membrane-targeting elements.
Nat Commun, 13, 2022
|
|
6TGX
 
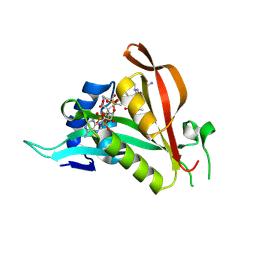 | | Crystal structure of Arabidopsis thaliana NAA60 in complex with a bisubstrate analogue | | Descriptor: | Acyl-CoA N-acyltransferases (NAT) superfamily protein, CARBOXYMETHYL COENZYME *A, MET-VAL-ASN-ALA | | Authors: | Layer, D, Kopp, J, Lapouge, K, Sinning, I. | | Deposit date: | 2019-11-18 | | Release date: | 2020-06-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The Arabidopsis N alpha -acetyltransferase NAA60 locates to the plasma membrane and is vital for the high salt stress response.
New Phytol., 228, 2020
|
|
2CIW
 
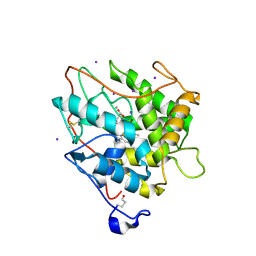 | | Chloroperoxidase iodide complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLOROPEROXIDASE, IODIDE ION, ... | | Authors: | Kuhnel, K, Blankenfeldt, W, Terner, J, Schlichting, I. | | Deposit date: | 2006-03-26 | | Release date: | 2006-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal Structures of Chloroperoxidase with its Bound Substrates and Complexed with Formate, Acetate, and Nitrate.
J.Biol.Chem., 281, 2006
|
|
5LN1
 
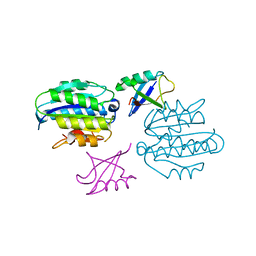 | | STRUCTURE OF UBIQUITYLATED-RPN10 FROM YEAST; | | Descriptor: | 26S proteasome regulatory subunit RPN10, Polyubiquitin-B | | Authors: | Keren-Kaplan, T, Attali, I, Levin-Kravets, O, Prag, G. | | Deposit date: | 2016-08-02 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Structure of ubiquitylated-Rpn10 provides insight into its autoregulation mechanism.
Nat Commun, 7, 2016
|
|
2NV0
 
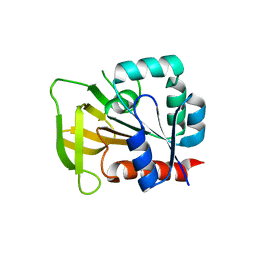 | |
2YM8
 
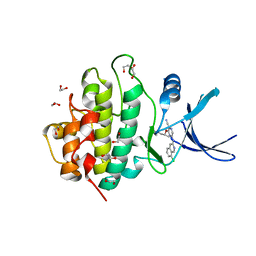 | | Crystal structure of checkpoint kinase 1 (Chk1) in complex with inhibitors | | Descriptor: | (R)-5-(8-CHLOROISOQUINOLIN-3-YLAMINO)-3-(1-(DIMETHYLAMINO)PROPAN-2-YLOXY)PYRAZINE-2-CARBONITRILE, 1,2-ETHANEDIOL, SERINE/THREONINE-PROTEIN KINASE CHK1 | | Authors: | Reader, J.C, Matthews, T.P, Klair, S, Cheung, K.M.J, Scanlon, J, Proisy, N, Addison, G, Ellard, J, Piton, N, Taylor, S, Cherry, M, Fisher, M, Boxall, K, Burns, S, Walton, M.I, Westwood, I.M, Hayes, A, Eve, P, Valenti, M, Brandon, A.H, Box, G, vanMontfort, R.L.M, Williams, D.H, Aherne, G.W, Raynaud, F.I, Eccles, S.A, Garrett, M.D, Collins, I. | | Deposit date: | 2011-06-06 | | Release date: | 2012-01-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure-Guided Evolution of Potent and Selective Chk1 Inhibitors Through Scaffold Morphing.
J.Med.Chem., 54, 2011
|
|
6IAH
 
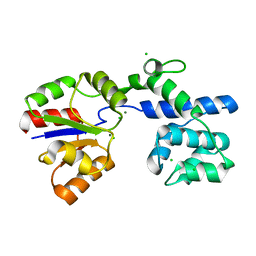 | | Phosphatase Tt82 from Thermococcus thioreducens | | Descriptor: | CHLORIDE ION, Hydrolase, MAGNESIUM ION | | Authors: | Havlickova, P, Brinsa, V, Brynda, J, Pachl, P, Prudnikova, T, Mesters, J.R, Kascakova, B, Kuty, M, Pusey, M.L, Ng, J.D, Rezacova, P, Smatanova, I.K. | | Deposit date: | 2018-11-26 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A novel structurally characterized haloacid dehalogenase superfamily phosphatase from Thermococcus thioreducens with diverse substrate specificity.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
8AX4
 
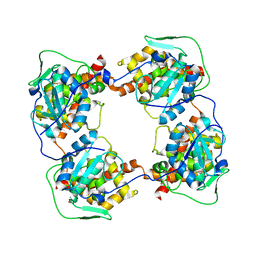 | |
8AXF
 
 | | Crystal structure of FMV N bound to 42-mer ssRNA | | Descriptor: | GLYCEROL, MAGNESIUM ION, Nucleocapsid protein, ... | | Authors: | Izhaki-Tavor, L, Yechezkel, I, Dessau, M. | | Deposit date: | 2022-08-31 | | Release date: | 2023-04-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | RNA Encapsulation Mode and Evolutionary Insights from the Crystal Structure of Emaravirus Nucleoprotein.
Microbiol Spectr, 11, 2023
|
|
6S1E
 
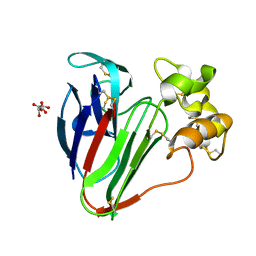 | | Structure of thaumatin determined at SwissFEL using native-SAD at 6.06 keV from all available diffraction patterns | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Nass, K, Cheng, R, Vera, L, Mozzanica, A, Redford, S, Ozerov, D, Basu, S, James, D, Knopp, G, Cirelli, C, Martiel, I, Casadei, C, Weinert, T, Nogly, P, Skopintsev, P, Usov, I, Leonarski, F, Geng, T, Rappas, M, Dore, A.S, Cooke, R, Nasrollahi Shirazi, S, Dworkowski, F, Sharpe, M, Olieric, N, Steinmetz, M.O, Schertler, G, Abela, R, Patthey, L, Schmitt, B, Hennig, M, Standfuss, J, Wang, M, Milne, J.C. | | Deposit date: | 2019-06-18 | | Release date: | 2020-07-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Advances in long-wavelength native phasing at X-ray free-electron lasers.
Iucrj, 7, 2020
|
|
5EDJ
 
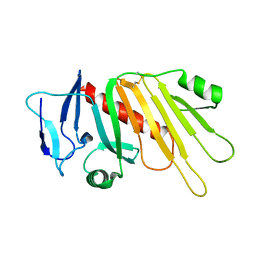 | | Crystal structure of the Neisseria meningitidis iron-regulated outer membrane lipoprotein FrpD | | Descriptor: | FrpC operon protein | | Authors: | Sviridova, E, Bumba, L, Rezacova, P, Sebo, P, Kuta Smatanova, I. | | Deposit date: | 2015-10-21 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of the interaction between the putative adhesion-involved and iron-regulated FrpD and FrpC proteins of Neisseria meningitidis.
Sci Rep, 7, 2017
|
|
8AYN
 
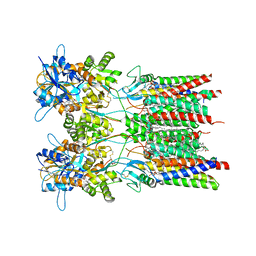 | | Resting state GluA1/A2 AMPA receptor in complex with TARP gamma 8 and ligand LY3130481 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 6-[(1~{S})-1-[1-[5-(2-hydroxyethyloxy)pyridin-2-yl]pyrazol-3-yl]ethyl]-3~{H}-1,3-benzothiazol-2-one, ... | | Authors: | Zhang, D, Lape, R, Shaikh, S, Kohegyi, B, Watson, J.F, Cais, O, Nakagawa, T, Greger, I.H. | | Deposit date: | 2022-09-02 | | Release date: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Modulatory mechanisms of TARP gamma 8-selective AMPA receptor therapeutics.
Nat Commun, 14, 2023
|
|
5LQV
 
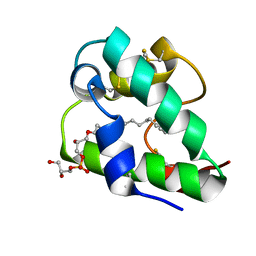 | | Spatial structure of the lentil lipid transfer protein in complex with anionic lysolipid LPPG | | Descriptor: | 1-MYRISTOYL-2-HYDROXY-SN-GLYCERO-3-[PHOSPHO-RAC-(1-GLYCEROL)], Non-specific lipid-transfer protein 2 | | Authors: | Mineev, K.S, Shenkarev, Z.O, Arseniev, A.S, Melnikova, D.N, Finkina, E.I, Ovchinnikova, T.V. | | Deposit date: | 2016-08-17 | | Release date: | 2017-06-28 | | Last modified: | 2019-05-08 | | Method: | SOLUTION NMR | | Cite: | Ligand Binding Properties of the Lentil Lipid Transfer Protein: Molecular Insight into the Possible Mechanism of Lipid Uptake.
Biochemistry, 56, 2017
|
|
6OO9
 
 | | Human CYP3A4 bound to a drug mibefradil | | Descriptor: | (1S,2S)-2-(2-{[3-(1H-benzimidazol-2-yl)propyl](methyl)amino}ethyl)-6-fluoro-1-(propan-2-yl)-1,2,3,4-tetrahydronaphthalen-2-yl methoxyacetate, 1,2-ETHANEDIOL, Cytochrome P450 3A4, ... | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2019-04-22 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Insights into the Interaction of Cytochrome P450 3A4 with Suicide Substrates: Mibefradil, Azamulin and 6',7'-Dihydroxybergamottin.
Int J Mol Sci, 20, 2019
|
|
6H9C
 
 | | Cryo-EM structure of archaeal extremophilic internal membrane-containing Haloarcula californiae icosahedral virus 1 (HCIV-1) at 3.74 Angstroms resolution. | | Descriptor: | (Half) GPS-II protein located underneath the two-tower capsomer sitting ON the icosahedral 2-fold axis., GPS-II protein located underneath the two-tower capsomer NOT sitting on the icosahedral 2-fold axis., GPS-III molecule located underneath the capsomer close to the icosahedral three-fold axis., ... | | Authors: | Abrescia, N.G, Santos-Perez, I, Charro, D. | | Deposit date: | 2018-08-03 | | Release date: | 2019-03-27 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Structural basis for assembly of vertical single beta-barrel viruses.
Nat Commun, 10, 2019
|
|
8A6E
 
 | | 100 picosecond light activated crystal structure of bovine rhodopsin in Lipidic Cubic Phase (SACLA) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gruhl, T, Weinert, T, Rodrigues, M.J, Milne, C.J, Ortolani, G, Nass, K, Nango, E, Sen, S, Johnson, P.J.M, Cirelli, C, Furrer, A, Mous, S, Skopintsev, P, James, D, Dworkowski, F, Baath, P, Kekilli, D, Oserov, D, Tanaka, R, Glover, H, Bacellar, C, Bruenle, S, Casadei, C.M, Diethelm, A.D, Gashi, D, Gotthard, G, Guixa-Gonzalez, R, Joti, Y, Kabanova, V, Knopp, G, Lesca, E, Ma, P, Martiel, I, Muehle, J, Owada, S, Pamula, F, Sarabi, D, Tejero, O, Tsai, C.J, Varma, N, Wach, A, Boutet, S, Tono, K, Nogly, P, Deupi, X, Iwata, S, Neutze, R, Standfuss, J, Schertler, G.F.X, Panneels, V. | | Deposit date: | 2022-06-17 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ultrafast structural changes direct the first molecular events of vision.
Nature, 615, 2023
|
|
5IUY
 
 | | Structural insights of the outer-membrane channel OprN | | Descriptor: | CHLORIDE ION, FORMIC ACID, Multidrug efflux outer membrane protein OprN, ... | | Authors: | Ntsogo, Y, Garnier, C, Phan, G, Monlezun, L, Benas, P, Broutin, I. | | Deposit date: | 2016-03-18 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Xenon for tunnelling analysis of the efflux pump component OprN.
PLoS ONE, 12, 2017
|
|
2CDE
 
 | | Structure and binding kinetics of three different human CD1d-alpha- Galactosylceramide specific T cell receptors - iNKT-TCR | | Descriptor: | INKT-TCR | | Authors: | Gadola, S.D, Koch, M, Marles-Wright, J, Lissin, N.M, Sheperd, D, Matulis, G, Harlos, K, Villiger, P.M, Stuart, D.I, Jakobsen, B.K, Cerundolo, V, Jones, E.Y. | | Deposit date: | 2006-01-23 | | Release date: | 2006-03-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structrue and Binding Kinetics of Three Different Human Cd1D-Alpha-Galactosylceramide-Specific T Cell Receptors
J.Exp.Med., 203, 2006
|
|
6OQZ
 
 | | Crystal structure of Glucose Isomerase from Non-merohedrally twinned crystals | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Sevvana, M, Ruf, M, Uson, I, Sheldrick, G.M, Herbst-Irmer, R. | | Deposit date: | 2019-04-29 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Non-merohedral twinning: from minerals to proteins.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
8A6D
 
 | | 10 picosecond light activated crystal structure of bovine rhodopsin in Lipidic Cubic Phase | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gruhl, T, Weinert, T, Rodrigues, M.J, Milne, C.J, Ortolani, G, Nass, K, Nango, E, Sen, S, Johnson, P.J.M, Cirelli, C, Furrer, A, Mous, S, Skopintsev, P, James, D, Dworkowski, F, Baath, P, Kekilli, D, Oserov, D, Tanaka, R, Glover, H, Bacellar, C, Bruenle, S, Casadei, C.M, Diethelm, A.D, Gashi, D, Gotthard, G, Guixa-Gonzalez, R, Joti, Y, Kabanova, V, Knopp, G, Lesca, E, Ma, P, Martiel, I, Muehle, J, Owada, S, Pamula, F, Sarabi, D, Tejero, O, Tsai, C.J, Varma, N, Wach, A, Boutet, S, Tono, K, Nogly, P, Deupi, X, Iwata, S, Neutze, R, Standfuss, J, Schertler, G.F.X, Panneels, V. | | Deposit date: | 2022-06-17 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ultrafast structural changes direct the first molecular events of vision.
Nature, 615, 2023
|
|
