7AR6
 
 | | Structure of apo SARS-CoV-2 Main Protease with large beta angle, space group C2. | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Andaleeb, H, Werner, N, Falke, S, Hinrichs, W, Alves Franca, B, Schwinzer, M, Brognaro, H, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Boger, J, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-10-23 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AWS
 
 | | Structure of SARS-CoV-2 Main Protease bound to TH-302. | | Descriptor: | 3C-like proteinase, 5-[[(2-bromoethylamino)-(ethylamino)phosphoryl]oxymethyl]-1-methyl-~{N},~{N}-bis(oxidanyl)imidazol-2-amine, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-11-09 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
8CBB
 
 | |
7ABU
 
 | | Structure of SARS-CoV-2 Main Protease bound to RS102895 | | Descriptor: | 1'-[2-[4-(trifluoromethyl)phenyl]ethyl]spiro[1~{H}-3,1-benzoxazine-4,4'-piperidine]-2-one, 3C-like proteinase, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Lane, T.J, Dunkel, I, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-09-08 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
3EJH
 
 | | Crystal Structure of the Fibronectin 8-9FnI Domain Pair in Complex with a Type-I Collagen Peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Collagen type-I a1 chain, Fibronectin, ... | | Authors: | Erat, M.C, Lowe, E.D, Campbell, I.D, Vakonakis, I. | | Deposit date: | 2008-09-18 | | Release date: | 2009-02-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification and structural analysis of type I collagen sites in complex with fibronectin fragments.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
7ADW
 
 | | Structure of SARS-CoV-2 Main Protease bound to 2,4'-Dimethylpropiophenone. | | Descriptor: | 2-methyl-1-(4-methylphenyl)propan-1-one, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-09-16 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AWT
 
 | | E. coli NADH quinone oxidoreductase hydrophilic arm | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Schimpf, J, Grishkovskaya, I, Haselbach, D, Friedrich, T. | | Deposit date: | 2020-11-09 | | Release date: | 2021-09-15 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Structure of the peripheral arm of a minimalistic respiratory complex I.
Structure, 30, 2022
|
|
6ZG3
 
 | | the structure of ECF PanT transporter in a complex with a nanobody | | Descriptor: | CA14381 nanobody, CITRIC ACID, Conserved hypothetical membrane protein, ... | | Authors: | Setyawati, I, Guskov, A, Slotboom, D.J. | | Deposit date: | 2020-06-18 | | Release date: | 2021-03-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | In vitro reconstitution of dynamically interacting integral membrane subunits of energy-coupling factor transporters.
Elife, 9, 2020
|
|
7NNP
 
 | | Rb-loaded cryo-EM structure of the E1-ATP KdpFABC complex. | | Descriptor: | CARDIOLIPIN, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Potassium-transporting ATPase ATP-binding subunit, ... | | Authors: | Silberberg, J.M, Corey, R.A, Hielkema, L, Stock, C, Stansfeld, P.J, Paulino, C, Haenelt, I. | | Deposit date: | 2021-02-25 | | Release date: | 2021-07-28 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Deciphering ion transport and ATPase coupling in the intersubunit tunnel of KdpFABC.
Nat Commun, 12, 2021
|
|
7NNL
 
 | | Cryo-EM structure of the KdpFABC complex in an E1-ATP conformation loaded with K+ | | Descriptor: | CARDIOLIPIN, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, POTASSIUM ION, ... | | Authors: | Silberberg, J.M, Corey, R.A, Hielkema, L, Stock, C, Stansfeld, P.J, Paulino, C, Haenelt, I. | | Deposit date: | 2021-02-25 | | Release date: | 2021-07-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Deciphering ion transport and ATPase coupling in the intersubunit tunnel of KdpFABC.
Nat Commun, 12, 2021
|
|
8AVQ
 
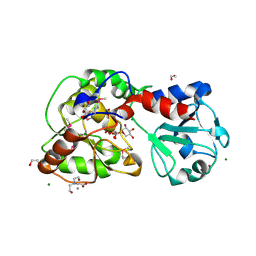 | | AO75L in Complex with UDP-Xylose | | Descriptor: | 1,2-ETHANEDIOL, AO75L, BICINE, ... | | Authors: | Laugeri, M.E, Speciale, I, Gimeno, A, Lin, S, Poveda, A, Lowary, T, Van Etten, J.L, Barbero, J.J, De Castro, C, Tonetti, M, Rojas, A.L. | | Deposit date: | 2022-08-26 | | Release date: | 2023-09-06 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Unveiling the GT114 family: Structural characterization of A075L, a glycosyltransferase from Paramecium bursaria chlorella virus-1 (PBCV-1).
Protein Sci., 33, 2024
|
|
6ZAK
 
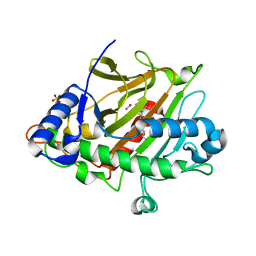 | | Room temperature XFEL Isopenicillin N synthase structure in complex with the Fe(IV)=O mimic VO and ACV. | | Descriptor: | Isopenicillin N synthase, L-D-(A-AMINOADIPOYL)-L-CYSTEINYL-D-VALINE, SULFATE ION, ... | | Authors: | Rabe, P, Kamps, J.J.A.G, Sutherlin, K, Pharm, C, McDonough, M.A, Leissing, T.M, Aller, P, Butryn, A, Linyard, J, Lang, P, Brem, J, Fuller, F.D, Batyuk, A, Hunter, M.S, Pettinati, I, Clifton, I.J, Alonso-Mori, R, Gul, S, Young, I, Kim, I, Bhowmick, A, ORiordan, L, Brewster, A.S, Claridge, T.D.W, Sauter, N.K, Yachandra, V, Yano, J, Kern, J.F, Orville, A.M, Schofield, C.J. | | Deposit date: | 2020-06-05 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Room temperature XFEL Isopenicillin N synthase structure in complex with the Fe(IV)=O mimic VO and ACV.
To Be Published
|
|
6WQR
 
 | |
8BQY
 
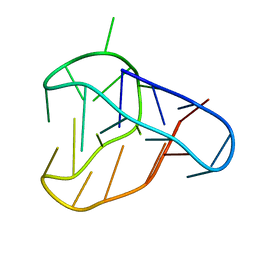 | | An i-motif domain able to undergo pH-dependent conformational transitions (acidic structure) | | Descriptor: | DNA (5'-D(*CP*(DNR)P*GP*TP*TP*(DNR)P*(DNR)P*GP*TP*TP*TP*TP*TP*CP*CP*GP*TP*TP*(DNR)P*CP*GP*T)-3') | | Authors: | Serrano-Chacon, I, Mir, B, Cupellini, L, Colizzi, F, Orozco, M, Escaja, N, Gonzalez, C. | | Deposit date: | 2022-11-22 | | Release date: | 2023-02-22 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | pH-Dependent Capping Interactions Induce Large-Scale Structural Transitions in i-Motifs.
J.Am.Chem.Soc., 145, 2023
|
|
8CCN
 
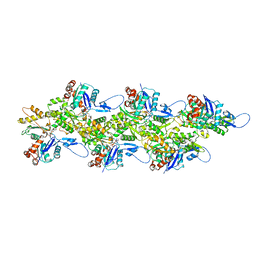 | | Filamentous actin II from Plasmodium falciparum | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-2, MAGNESIUM ION | | Authors: | Kursula, I, Lopez, A.J. | | Deposit date: | 2023-01-27 | | Release date: | 2023-02-22 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure and function of Plasmodium actin II in the parasite mosquito stages.
Plos Pathog., 19, 2023
|
|
8BV6
 
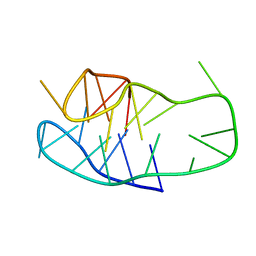 | | An i-motif domain able to undergo pH-dependent conformational transitions (neutral structure) | | Descriptor: | DNA (5'-D(*CP*(DNR)P*GP*TP*TP*CP*(DNR)P*GP*TP*TP*TP*TP*TP*CP*CP*GP*TP*TP*CP*CP*GP*T)-3') | | Authors: | Serrano-Chacon, I, Mir, B, Cupellini, L, Colizzi, F, Orozco, M, Escaja, N, Gonzalez, C. | | Deposit date: | 2022-12-01 | | Release date: | 2023-02-22 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | pH-Dependent Capping Interactions Induce Large-Scale Structural Transitions in i-Motifs.
J.Am.Chem.Soc., 145, 2023
|
|
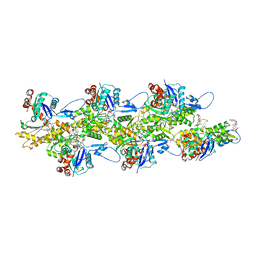
8CCO
 
 | |
6ZZ8
 
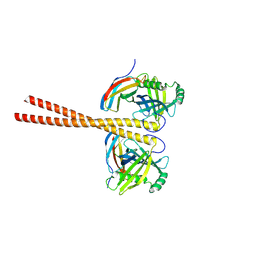 | | MB_CRS6-15 bound to CrSAS-6_6HR | | Descriptor: | Centriole protein, Protein B | | Authors: | Hatzopoulos, G.N, Kukenshoner, T, Banterle, N, Favez, T, Fluckiger, I, Hantschel, O, Gonczy, P. | | Deposit date: | 2020-08-04 | | Release date: | 2021-07-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.73 Å) | | Cite: | Tuning SAS-6 architecture with monobodies impairs distinct steps of centriole assembly.
Nat Commun, 12, 2021
|
|
1A1N
 
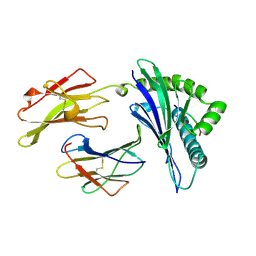 | | MHC CLASS I MOLECULE B*3501 COMPLEXED WITH PEPTIDE VPLRPMTY FROM THE NEF PROTEIN (75-82) OF HIV1 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, BW-53 B*5301 alpha chain, ... | | Authors: | Smith, K.J, Reid, S.W, Stuart, D.I, Mcmichael, A.J, Jones, E.Y, Bell, J.I. | | Deposit date: | 1997-12-11 | | Release date: | 1998-04-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An altered position of the alpha 2 helix of MHC class I is revealed by the crystal structure of HLA-B*3501.
Immunity, 4, 1996
|
|
1A1O
 
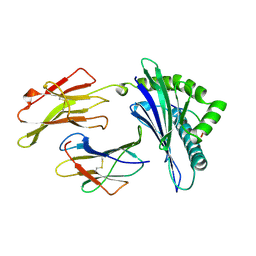 | | MHC CLASS I MOLECULE B*5301 COMPLEXED WITH PEPTIDE LS6 (KPIVQYDNF) FROM THE MALARIA PARASITE P. FALCIPARUM | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, BW-53 B*5301 alpha chain, ... | | Authors: | Smith, K.J, Reid, S.W, Harlos, K, Mcmichael, A.J, Stuart, D.I, Bell, J.I, Jones, E.Y. | | Deposit date: | 1997-12-11 | | Release date: | 1998-04-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bound water structure and polymorphic amino acids act together to allow the binding of different peptides to MHC class I HLA-B53.
Immunity, 4, 1996
|
|
8C9G
 
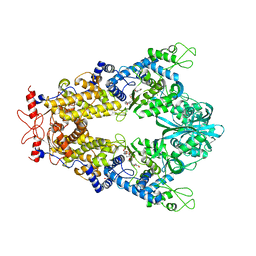 | | Priestia megaterium mupirocin-sensitive isoleucyl-tRNA synthetase 1 complexed with mupirocin | | Descriptor: | CHLORIDE ION, GLYCEROL, Isoleucine--tRNA ligase, ... | | Authors: | Brkic, A, Leibundgut, M, Jablonska, J, Zanki, V, Car, Z, Petrovic Perokovic, V, Ban, N, Gruic-Sovulj, I. | | Deposit date: | 2023-01-22 | | Release date: | 2023-08-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Antibiotic hyper-resistance in a class I aminoacyl-tRNA synthetase with altered active site signature motif.
Nat Commun, 14, 2023
|
|
8C9D
 
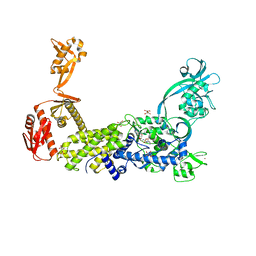 | | Priestia megaterium W130Q mutant of type 2 isoleucyl-tRNA synthetase complexed with an isoleucyl-adenylate analogue | | Descriptor: | D(-)-TARTARIC ACID, Isoleucine--tRNA ligase, N-[ISOLEUCINYL]-N'-[ADENOSYL]-DIAMINOSUFONE, ... | | Authors: | Brkic, A, Leibundgut, M, Jablonska, J, Zanki, V, Car, Z, Petrovic Perokovic, V, Ban, N, Gruic-Sovulj, I. | | Deposit date: | 2023-01-21 | | Release date: | 2023-08-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Antibiotic hyper-resistance in a class I aminoacyl-tRNA synthetase with altered active site signature motif.
Nat Commun, 14, 2023
|
|
8C8V
 
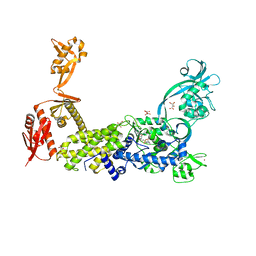 | | Priestia megaterium mupirocin-resistant isoleucyl-tRNA synthetase 2 with a fully-resolved C-terminal tRNA-binding domain complexed with an isoleucyl-adenylate analogue | | Descriptor: | D(-)-TARTARIC ACID, Isoleucine--tRNA ligase, N-[ISOLEUCINYL]-N'-[ADENOSYL]-DIAMINOSUFONE, ... | | Authors: | Brkic, A, Leibundgut, M, Jablonska, J, Zanki, V, Car, Z, Petrovic Perokovic, V, Ban, N, Gruic-Sovulj, I. | | Deposit date: | 2023-01-21 | | Release date: | 2023-08-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Antibiotic hyper-resistance in a class I aminoacyl-tRNA synthetase with altered active site signature motif.
Nat Commun, 14, 2023
|
|
8C9E
 
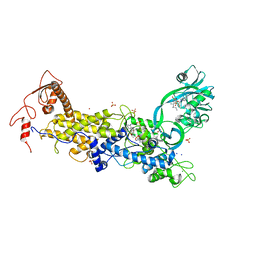 | | Priestia megaterium mupirocin-sensitive isoleucyl-tRNA synthetase 1 complexed with an isoleucyl-adenylate analogue | | Descriptor: | CHLORIDE ION, Isoleucine--tRNA ligase, LITHIUM ION, ... | | Authors: | Brkic, A, Leibundgut, M, Jablonska, J, Zanki, V, Car, Z, Petrovic Perokovic, V, Ban, N, Gruic-Sovulj, I. | | Deposit date: | 2023-01-21 | | Release date: | 2023-08-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Antibiotic hyper-resistance in a class I aminoacyl-tRNA synthetase with altered active site signature motif.
Nat Commun, 14, 2023
|
|
7ANS
 
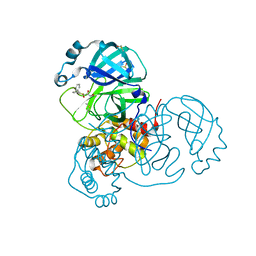 | | Structure of SARS-CoV-2 Main Protease bound to Adrafinil. | | Descriptor: | 2-[(diphenylmethyl)-oxidanyl-$l^{3}-sulfanyl]-~{N}-oxidanyl-ethanamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Ewert, W, Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-10-12 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
