3SPK
 
 | | Tipranavir in Complex with a Human Immunodeficiency Virus Type 1 Protease Variant | | Descriptor: | HIV-1 protease, N-(3-{(1R)-1-[(6R)-4-HYDROXY-2-OXO-6-PHENETHYL-6-PROPYL-5,6-DIHYDRO-2H-PYRAN-3-YL]PROPYL}PHENYL)-5-(TRIFLUOROMETHYL)-2-PYRIDINESULFONAMIDE | | Authors: | Wang, Y, Liu, Z, Brunzelle, J.S, Kovari, I.A, Kovari, L.C. | | Deposit date: | 2011-07-01 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | The higher barrier of darunavir and tipranavir resistance for HIV-1 protease.
Biochem.Biophys.Res.Commun., 412, 2011
|
|
3EZV
 
 | | CDK-2 with indazole inhibitor 9 bound at its active site | | Descriptor: | 4-{3-[7-(4-methylpiperazin-1-yl)-1H-benzimidazol-2-yl]-1H-indazol-6-yl}aniline, Cell division protein kinase 2 | | Authors: | Kiefer, J.R, Day, J.E, Caspers, N.L, Mathis, K.J, Kretzmer, K.K, Weinberg, R.A, Reitz, B.A, Stegeman, R.A, Trujillo, J.I, Huang, W, Thorarensen, A, Xing, L, Wrightstone, A, Christine, L, Compton, R, Li, X. | | Deposit date: | 2008-10-23 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | 2-(6-Phenyl-1H-indazol-3-yl)-1H-benzo[d]imidazoles: Design and synthesis of a potent and isoform selective PKC-zeta inhibitor
Bioorg.Med.Chem.Lett., 19, 2009
|
|
2OUG
 
 | |
5B0D
 
 | | Polyketide cyclase OAC from Cannabis sativa, Y27W mutant | | Descriptor: | Olivetolic acid cyclase | | Authors: | Yang, X, Matsui, T, Mori, T, Abe, I, Morita, H. | | Deposit date: | 2015-10-28 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural basis for olivetolic acid formation by a polyketide cyclase from Cannabis sativa
Febs J., 283, 2016
|
|
1FXK
 
 | |
5APD
 
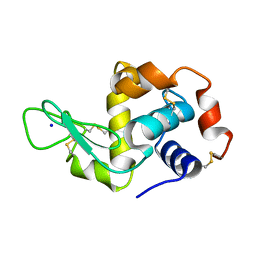 | | Hen Egg White Lysozyme not illuminated with 0.4THz radiation | | Descriptor: | LYSOZYME C, SODIUM ION | | Authors: | Lundholm, I, Rodilla, H, Wahlgren, W.Y, Duelli, A, Bourenkov, G, Vukusic, J, Friedman, R, Stake, J, Schneider, T, Katona, G. | | Deposit date: | 2015-09-15 | | Release date: | 2016-01-13 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Terahertz Radiation Induces Non-Thermal Structural Changes Associated with Frohlich Condensation in a Protein Crystal
Struct.Dyn., 2, 2015
|
|
3FE0
 
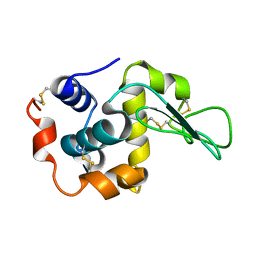 | | X-ray crystal structure of wild type human lysozyme in D2O | | Descriptor: | Lysozyme C | | Authors: | Chiba-Kamoshida, K, Matsui, T, Chatake, T, Ohhara, T, Ostermann, A, Tanaka, I, Yutani, K, Niimura, N. | | Deposit date: | 2008-11-27 | | Release date: | 2009-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Site-specific softening of peptide bonds by localized deuterium observed by neutron crystallography of human lysozyme hydrogen
To be Published
|
|
5APM
 
 | | Multiple capsid-stabilizing protein-RNA and protein-protein interactions revealed in a high-resolution structure of an emerging picornavirus causing neonatal sepsis | | Descriptor: | VP0, VP1, VP3 | | Authors: | Shakeel, S, Westerhuis, B.M, Domanska, A, Koning, R.I, Matadeen, R, Koster, A.J, Bakker, A.Q, Beaumont, T, Wolthers, K.C, Butcher, S.J. | | Deposit date: | 2015-09-17 | | Release date: | 2016-07-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Multiple Capsid-Stabilizing Interactions Revealed in a High-Resolution Structure of an Emerging Picornavirus Causing Neonatal Sepsis
Nat.Commun., 7, 2016
|
|
1G1E
 
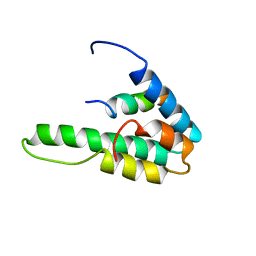 | | NMR STRUCTURE OF THE HUMAN MAD1 TRANSREPRESSION DOMAIN SID IN COMPLEX WITH MAMMALIAN SIN3A PAH2 DOMAIN | | Descriptor: | MAD1 PROTEIN, SIN3A | | Authors: | Brubaker, K, Cowley, S.M, Huang, K, Eisenman, R.N, Radhakrishnan, I. | | Deposit date: | 2000-10-11 | | Release date: | 2000-12-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the interacting domains of the Mad-Sin3 complex: implications for recruitment of a chromatin-modifying complex.
Cell(Cambridge,Mass.), 103, 2000
|
|
5APA
 
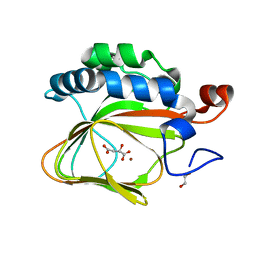 | | Crystal structure of human aspartate beta-hydroxylase isoform a | | Descriptor: | (2S)-2-hydroxybutanedioic acid, 1,2-ETHANEDIOL, ASPARTYL/ASPARAGINYL BETA-HYDROXYLASE, ... | | Authors: | Krojer, T, Kochan, G, Pfeffer, I, McDonough, M.A, Pilka, E, Hozjan, V, Allerston, C, Muniz, J.R, Chaikuad, A, Gileadi, O, Kavanagh, K, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Oppermann, U. | | Deposit date: | 2015-09-15 | | Release date: | 2015-09-23 | | Last modified: | 2019-11-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Aspartate/asparagine-beta-hydroxylase crystal structures reveal an unexpected epidermal growth factor-like domain substrate disulfide pattern.
Nat Commun, 10, 2019
|
|
5AWN
 
 | |
1QRK
 
 | | HUMAN FACTOR XIII WITH STRONTIUM BOUND IN THE ION SITE | | Descriptor: | PROTEIN (COAGULATION FACTOR XIII), STRONTIUM ION | | Authors: | Fox, B.A, Yee, V.C, Pederson, L.C, Le Trong, I, Bishop, P.D, Stenkamp, R.E, Teller, D.C. | | Deposit date: | 1999-06-14 | | Release date: | 1999-07-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of the calcium binding site and a novel ytterbium site in blood coagulation factor XIII by x-ray crystallography.
J.Biol.Chem., 274, 1999
|
|
1FW7
 
 | |
2OXC
 
 | | Human DEAD-box RNA helicase DDX20, DEAD domain in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Probable ATP-dependent RNA helicase DDX20 | | Authors: | Karlberg, T, Ogg, D, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A, Flodin, S, Flores, A, Graslund, S, Hallberg, B.M, Hammarstrom, M, Hogbom, M, Johansson, I, Kotenyova, T, Lehtio, L, Moche, M, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Stenmark, P, Sundstrom, M, Thorsell, A.G, Van Den Berg, S, Weigelt, J, Holmberg-Schiavone, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-02-20 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Human DEAD-box RNA helicase DDX20
To be Published
|
|
5ANG
 
 | | Crystal structure of CDK2 in complex with 7-hydroxy-4-(morpholinomethyl)chromen-2-one processed with the CrystalDirect automated mounting and cryo-cooling technology | | Descriptor: | 7-HYDROXY-4-(MORPHOLINOMETHYL)CHROMEN-2-ONE, CYCLIN-DEPENDENT KINASE 2 | | Authors: | Zander, U, Hoffmann, G, Mathieu, M, Marquette, J.-P, Cornaciu, I, Cipriani, F, Marquez, J.A. | | Deposit date: | 2015-09-07 | | Release date: | 2016-04-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Automated Harvesting and Processing of Protein Crystals Through Laser Photoablation.
Acta Crystallogr.,Sect.D, 72, 2016
|
|
3FB3
 
 | | Crystal Structure of Trypanosoma Brucei Acetyltransferase, Tb11.01.2886 | | Descriptor: | N-acetyltransferase | | Authors: | Wernimont, A.K, Marino, K, Zhang, A.Z, Ma, D, Lin, Y.H, MacKenzie, F, Kozieradzki, I, Cossar, D, Zhao, Y, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Ferguson, M.A.J, Hui, R, Qiu, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-11-18 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of Trypanosoma Brucei Acetyltransferase, Tb11.01.2886
TO BE PUBLISHED
|
|
3FE5
 
 | | Crystal structure of 3-hydroxyanthranilate 3,4-dioxygenase from bovine kidney | | Descriptor: | 3-hydroxyanthranilate 3,4-dioxygenase, FE (III) ION | | Authors: | Dilovic, I, Gliubich, F, Malpeli, G, Zanotti, G, Matkovic-Calogovic, D. | | Deposit date: | 2008-11-27 | | Release date: | 2009-06-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal structure of bovine 3-hydroxyanthranilate 3,4-dioxygenase.
Biopolymers, 2009
|
|
1GC5
 
 | | CRYSTAL STRUCTURE OF A NOVEL ADP-DEPENDENT GLUCOKINASE FROM THERMOCOCCUS LITORALIS | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADP-DEPENDENT GLUCOKINASE | | Authors: | Ito, S, Fushinobu, S, Yoshioka, I, Koga, S, Matsuzawa, H, Wakagi, T. | | Deposit date: | 2000-07-20 | | Release date: | 2001-07-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for the ADP-Specificity of a Novel Glucokinase from a Hyperthermophilic Archaeon
Structure, 9, 2001
|
|
1QCJ
 
 | | LOW TEMPERATURE COMPLEX OF POKEWEED ANTIVIRAL PROTEIN WITH PTEORIC ACID | | Descriptor: | 2-AMINO-6-[(4-CARBOXY-PHENYLAMINO)-METHYL]-4-HYDROXY-PTERIDIN-1-IUM, POKEWEED ANTIVIRAL PROTEIN | | Authors: | Kurinov, I.V, Myers, D.E, Irvin, J.D, Uckun, F.M. | | Deposit date: | 1999-05-05 | | Release date: | 1999-09-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray crystallographic analysis of the structural basis for the interactions of pokeweed antiviral protein with its active site inhibitor and ribosomal RNA substrate analogs.
Protein Sci., 8, 1999
|
|
2OOM
 
 | | NMR structure of a kissing complex formed between the TAR RNA element of HIV-1 and a LNA/RNA aptamer | | Descriptor: | RNA 16-mer, TAR RNA element of HIV-1 | | Authors: | Lebars, I, Richard, T, Di Primo, C, Toulme, J.J. | | Deposit date: | 2007-01-26 | | Release date: | 2008-01-29 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a kissing complex formed between the TAR RNA element of HIV-1 and a LNA-modified aptamer
Nucleic Acids Res., 35, 2007
|
|
2OPV
 
 | |
1GCW
 
 | | CO form hemoglobin from mustelus griseus | | Descriptor: | CARBON MONOXIDE, PROTEIN (HEMOGLOBIN), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Naoi, Y, Chong, K.T, Yoshimatsu, K, Miyazaki, G, Tame, J.R.H, Park, S.Y, Adachi, S.I, Morimoto, H. | | Deposit date: | 2000-08-08 | | Release date: | 2000-09-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The functional similarity and structural diversity of human and cartilaginous fish hemoglobins.
J.Mol.Biol., 307, 2001
|
|
1QZM
 
 | | alpha-domain of ATPase | | Descriptor: | ATP-dependent protease La | | Authors: | Botos, I, Melnikov, E.E, Cherry, S, Khalatova, A.G, Rasulova, F.S, Tropea, J.E, Maurizi, M.R, Rotanova, T.V, Gustchina, A, Wlodawer, A. | | Deposit date: | 2003-09-17 | | Release date: | 2004-05-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the AAA+ alpha domain of E. coli Lon protease at 1.9A resolution.
J.Struct.Biol., 146
|
|
1QZL
 
 | | GCATGCT + Cobalt | | Descriptor: | 5'-D(*GP*CP*AP*TP*GP*CP*T)-3', COBALT (II) ION | | Authors: | Cardin, C.J, Gan, Y, Thorpe, J.H, Teixeira, S.C.M, Gale, B.C, Moraes, M.I.A. | | Deposit date: | 2003-09-17 | | Release date: | 2003-10-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Metal Ion Distribution and Stabilisation of the DNA Quadruplex Structure Formed by d(GCATGCT)
To be published
|
|
1QZW
 
 | | Crystal structure of the complete core of archaeal SRP and implications for inter-domain communication | | Descriptor: | 7S RNA, Signal recognition 54 kDa protein | | Authors: | Rosendal, K.R, Wild, K, Montoya, G, Sinning, I. | | Deposit date: | 2003-09-18 | | Release date: | 2003-11-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Crystal structure of the complete core of archaeal signal recognition particle and implications for interdomain communication
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
