1UB9
 
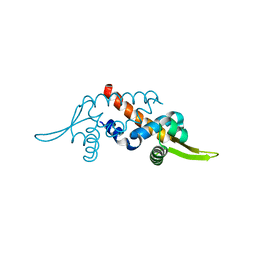 | | Structure of the transcriptional regulator homologue protein from Pyrococcus horikoshii OT3 | | Descriptor: | Hypothetical protein PH1061 | | Authors: | Okada, U, Sakai, N, Tajika, Y, Yao, M, Watanabe, N, Tanaka, I. | | Deposit date: | 2003-04-03 | | Release date: | 2004-05-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural analysis of the transcriptional regulator homolog protein from Pyrococcus horikoshii OT3.
Proteins, 63, 2006
|
|
1UCC
 
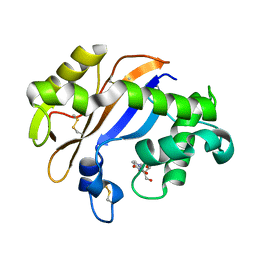 | | Crystal structure of the Ribonuclease MC1 from bitter gourd seeds complexed with 3'-UMP. | | Descriptor: | 3'-URIDINEMONOPHOSPHATE, Ribonuclease MC | | Authors: | Suzuki, A, Yao, M, Tanaka, I, Numata, T, Kikukawa, S, Yamasaki, N, Kimura, M. | | Deposit date: | 2003-04-10 | | Release date: | 2003-04-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structures of the ribonuclease MC1 from bitter gourd seeds, complexed with 2'-UMP or 3'-UMP, reveal structural basis for uridine specificity
Biochem.Biophys.Res.Commun., 275, 2000
|
|
1UEG
 
 | |
1U9L
 
 | | Structural basis for a NusA- protein N interaction | | Descriptor: | GOLD ION, Lambda N, Transcription elongation protein nusA | | Authors: | Bonin, I, Muehlberger, R, Bourenkov, G.P, Huber, R, Bacher, A, Richter, G, Wahl, M.C. | | Deposit date: | 2004-08-10 | | Release date: | 2004-08-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the interaction of Escherichia coli NusA with protein N of phage lambda
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
9M26
 
 | | Crystal structure of Enterobacter cloacae YcdY, a member of the redox enzyme maturation protein family | | Descriptor: | CALCIUM ION, Cytoplasmic chaperone TorD family protein | | Authors: | Choi, H.J, Lee, S.J, Kim, J.H, Yoon, S.I. | | Deposit date: | 2025-02-27 | | Release date: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of YcdY, a member of the redox-enzyme maturation protein family.
Acta Crystallogr.,Sect.F, 81, 2025
|
|
9QXW
 
 | |
9UJ2
 
 | | 14-3-3 zeta chimera with the S202R peptide of SARS-CoV-2 N (residues 200-213) | | Descriptor: | 1,2-ETHANEDIOL, 14-3-3 protein zeta/delta,Peptide from Nucleoprotein, DI(HYDROXYETHYL)ETHER | | Authors: | Boyko, K.M, Matyuta, I.O, Minyaev, M.E, Perfilova, K.V, Sluchanko, N.N. | | Deposit date: | 2025-04-16 | | Release date: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution structure reveals enhanced 14-3-3 binding by a mutant SARS-CoV-2 nucleoprotein variant with improved replicative fitness.
Biochem.Biophys.Res.Commun., 767, 2025
|
|
9L2N
 
 | | Crystal structure of Cytochalasin D bound to a filamentous conformation actin | | Descriptor: | (3S,3aR,4S,6S,6aR,7E,10S,12R,13E,15R,15aR)-3-benzyl-6,12-dihydroxy-4,10,12-trimethyl-5-methylidene-1,11-dioxo-2,3,3a,4,5,6,6a,9,10,11,12,15-dodecahydro-1H-cycloundeca[d]isoindol-15-yl acetate, 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Takeda, S, Maeda, Y, Fujiwara, I. | | Deposit date: | 2024-12-17 | | Release date: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Microscopic and structural observations of actin filament capping and severing by cytochalasin D
Proc.Natl.Acad.Sci.USA, 2025
|
|
2I66
 
 | | Structural Basis for the Mechanistic Understanding Human CD38 Controlled Multiple Catalysis | | Descriptor: | ADP-ribosyl cyclase 1, [(2R,3R,4R,5R)-5-(2-AMINO-6-OXO-1,6-DIHYDRO-9H-PURIN-9-YL)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHYL [(2R,3S,4R,5S)-3,4,5-TRIHYDROXYTETRAHYDROFURAN-2-YL]METHYL DIHYDROGEN DIPHOSPHATE, [(2R,3R,4R,5R)-5-(2-AMINO-6-OXO-1,6-DIHYDRO-9H-PURIN-9-YL)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHYL [(2R,3S,4S)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHYL DIHYDROGEN DIPHOSPHATE | | Authors: | Liu, Q, Kriksunov, I.A, Graeff, R, Munshi, C, Lee, H.C, Hao, Q. | | Deposit date: | 2006-08-28 | | Release date: | 2006-09-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the mechanistic understanding of human CD38-controlled multiple catalysis.
J.Biol.Chem., 281, 2006
|
|
6KYC
 
 | | Structure of the S207A mutant of Clostridium difficile sortase B | | Descriptor: | Putative peptidase C60B, sortase B | | Authors: | Kang, C.Y, Huang, I.H, Wu, T.Y, Chang, J.C, Hsiao, Y.Y, Cheng, C.H, Tsai, W.J, Hsu, K.C, Wang, S.Y. | | Deposit date: | 2019-09-17 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Functional analysis ofClostridium difficilesortase B reveals key residues for catalytic activity and substrate specificity.
J.Biol.Chem., 295, 2020
|
|
2I65
 
 | | Structural Basis for the Mechanistic Understanding Human CD38 Controlled Multiple Catalysis | | Descriptor: | ADP-ribosyl cyclase 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Liu, Q, Kriksunov, I.A, Graeff, R, Munshi, C, Lee, H.C, Hao, Q. | | Deposit date: | 2006-08-28 | | Release date: | 2006-09-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the mechanistic understanding of human CD38-controlled multiple catalysis.
J.Biol.Chem., 281, 2006
|
|
9OAR
 
 | |
9OVI
 
 | | Crystal Structure of SH3-like_bac-type domain (79-145) of Conserved domain protein GBAA_2967 from Bacillus anthracis Ames ancestor | | Descriptor: | Conserved domain protein, FORMIC ACID, MAGNESIUM ION | | Authors: | Minasov, G, Dubrovska, I, Winsor, J, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2025-05-30 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of SH3-like_bac-type domain (79-145) of Conserved domain protein GBAA_2967 from Bacillus anthracis Ames ancestor.
To Be Published
|
|
9OAO
 
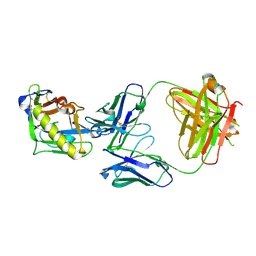 | |
9M5N
 
 | | Plasmodium vivax aspartyl-tRNA synthetase in complex with aspartyl-adenylate (Asp-AMP) Complex. | | Descriptor: | ACETATE ION, ASPARTYL-ADENOSINE-5'-MONOPHOSPHATE, GLYCEROL, ... | | Authors: | Sharma, V.K, Manickam, Y, Sharma, A, Bagale, S, Pradeepkumar, P.I. | | Deposit date: | 2025-03-06 | | Release date: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Natural product-mediated reaction hijacking mechanism validates Plasmodium aspartyl-tRNA synthetase as an antimalarial drug target
To Be Published
|
|
9OAS
 
 | |
9M5O
 
 | | Plasmodium vivax aspartyl-tRNA synthetase in complex with aspartyl sulfamoyl adenosine (Asp-AMS) Complex | | Descriptor: | 5'-O-(L-alpha-aspartylsulfamoyl)adenosine, MAGNESIUM ION, aspartate--tRNA ligase | | Authors: | Sharma, V.K, Manickam, Y, Sharma, A, Bagale, S, Pradeepkumar, P.I. | | Deposit date: | 2025-03-06 | | Release date: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (1.839 Å) | | Cite: | Natural product-mediated reaction hijacking mechanism validates Plasmodium aspartyl-tRNA synthetase as an antimalarial drug target
To Be Published
|
|
9QXY
 
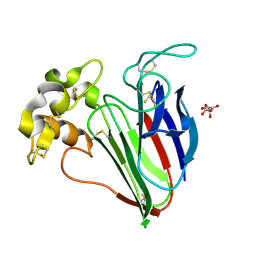 | |
9NR2
 
 | | Crystal structure of H5 hemagglutinin from the influenza virus A/black swan/Akita/1/2016 with LSTa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, Hemagglutinin HA2, ... | | Authors: | Lin, T.H, Zhu, Y, Wilson, I.A. | | Deposit date: | 2025-03-13 | | Release date: | 2025-04-23 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | The Q226L mutation can convert a highly pathogenic H5 2.3.4.4e virus to bind human-type receptors.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9M5M
 
 | | Crystal structure of Plasmodium vivax aspartyl-tRNA synthetase (PvDRS) | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, aspartate--tRNA ligase | | Authors: | Sharma, V.K, Manickam, Y, Bagale, S, Pradeepkumar, P.I, Sharma, A. | | Deposit date: | 2025-03-06 | | Release date: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Natural product-mediated reaction hijacking mechanism validates Plasmodium aspartyl-tRNA synthetase as an antimalarial drug target
To Be Published
|
|
2IBN
 
 | | Crystal structure of Human myo-Inositol Oxygenase (MIOX) | | Descriptor: | (2S,3R,4R,5S,6S)-2,3,4,5,6-PENTAHYDROXYCYCLOHEXANONE, CYSTEINE, FE (III) ION, ... | | Authors: | Hallberg, B.M, Busam, R.D, Arrowsmith, C, Berglund, H, Collins, R, Edwards, A, Ehn, M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hogbom, M, Holmberg-Schiavone, L, Johansson, I, Karlberg, T, Kotenyova, T, Nilsson-Ehle, P, Nordlund, P, Nyman, T, Ogg, D, Sagemark, J, Stenmark, P, Sundstrom, M, Uppenberg, J, Van Den Berg, S, Weigelt, J, Thorsell, A.G, Persson, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-09-11 | | Release date: | 2006-10-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Biophysical Characterization of Human myo-Inositol Oxygenase
J.Biol.Chem., 283, 2008
|
|
7F1D
 
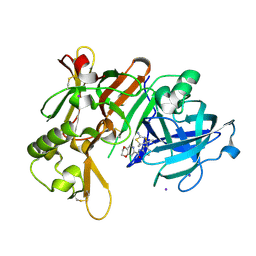 | | Crystal Structure of BACE1 in complex with N-{3-[(4R,5R,6R)-2-amino-5-fluoro-4,6-dimethyl-5,6-dihydro-4H-1,3-thiazin-4-yl]-4-fluorophenyl}-2H,3H-[1,4]dioxino[2,3-c]pyridine-7-carboxamide | | Descriptor: | Beta-secretase 1, IODIDE ION, N-[3-[(4R,5R,6R)-2-azanyl-5-fluoranyl-4,6-dimethyl-5,6-dihydro-1,3-thiazin-4-yl]-4-fluoranyl-phenyl]-2,3-dihydro-[1,4]dioxino[2,3-c]pyridine-7-carboxamide | | Authors: | Ueno, T, Matsuoka, E, Asada, N, Yamamoto, S, Kanegawa, N, Ito, M, Ito, H, Moechars, D, Rombouts, F.J.R, Gijsen, H.J.M, Kusakabe, K.I. | | Deposit date: | 2021-06-09 | | Release date: | 2022-02-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of Extremely Selective Fused Pyridine-Derived beta-Site Amyloid Precursor Protein-Cleaving Enzyme (BACE1) Inhibitors with High In Vivo Efficacy through 10s Loop Interactions.
J.Med.Chem., 64, 2021
|
|
2I5D
 
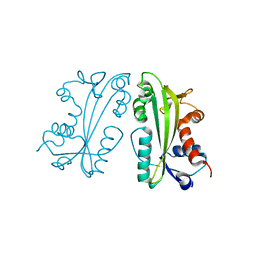 | | Crystal Structure of Human Inosine Triphosphate Pyrophosphatase | | Descriptor: | inosine triphosphate pyrophosphohydrolase | | Authors: | Porta, J.C, Kozmin, S.G, Pavlov, Y.I, Borgstahl, G.E.O. | | Deposit date: | 2006-08-24 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structure of the orthorhombic form of human inosine triphosphate pyrophosphatase.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
5JXV
 
 | | Solid-state MAS NMR structure of immunoglobulin beta 1 binding domain of protein G (GB1) | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Andreas, L.B, Jaudzems, K, Stanek, J, Lalli, D, Bertarello, A, Le Marchand, T, Cala-De Paepe, D, Kotelovica, S, Akopjana, I, Knott, B, Wegner, S, Engelke, F, Lesage, A, Emsley, L, Tars, K, Herrmann, T, Pintacuda, G. | | Deposit date: | 2016-05-13 | | Release date: | 2016-08-10 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | Structure of fully protonated proteins by proton-detected magic-angle spinning NMR.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5JZR
 
 | | Solid-state MAS NMR structure of Acinetobacter phage 205 (AP205) coat protein in assembled capsid particles | | Descriptor: | Coat protein | | Authors: | Jaudzems, K, Andreas, L.B, Stanek, J, Lalli, D, Bertarello, A, Le Marchand, T, Cala-De Paepe, D, Kotelovica, S, Akopjana, I, Knott, B, Wegner, S, Engelke, F, Lesage, A, Emsley, L, Tars, K, Herrmann, T, Pintacuda, G. | | Deposit date: | 2016-05-17 | | Release date: | 2016-08-10 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | Structure of fully protonated proteins by proton-detected magic-angle spinning NMR.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
