1BQG
 
 | | THE STRUCTURE OF THE D-GLUCARATE DEHYDRATASE PROTEIN FROM PSEUDOMONAS PUTIDA | | Descriptor: | D-GLUCARATE DEHYDRATASE | | Authors: | Gulick, A.M, Palmer, D.R.J, Babbitt, P.C, Gerlt, J.A, Rayment, I. | | Deposit date: | 1998-08-15 | | Release date: | 1999-05-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: crystal structure of (D)-glucarate dehydratase from Pseudomonas putida.
Biochemistry, 37, 1998
|
|
6S0O
 
 | | Crystal Structure of Two-Domain Laccase from Streptomyces griseoflavus produced at 0.25 mM copper sulfate in growth medium | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, GLYCEROL, ... | | Authors: | Gabdulkhakov, A.G, Tishchenko, T.V, Kolyadenko, I.A. | | Deposit date: | 2019-06-17 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Investigations of Accessibility of T2/T3 Copper Center of Two-Domain Laccase fromStreptomyces griseoflavusAc-993.
Int J Mol Sci, 20, 2019
|
|
2BR7
 
 | | Crystal Structure of Acetylcholine-binding Protein (AChBP) from Aplysia californica in complex with HEPES | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, SOLUBLE ACETYLCHOLINE RECEPTOR | | Authors: | Celie, P.H.N, Kasheverov, I.E, Mordvintsev, D.Y, Hogg, R.C, Van Nierop, P, Van Elk, R, Van Rossum-Fikkert, S.E, Zhmak, M.N, Bertrand, D, Tsetlin, V, Sixma, T.K, Smit, A.B. | | Deposit date: | 2005-05-03 | | Release date: | 2005-06-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of Nicotinic Acetylcholine Receptor Homolog Achbp in Complex with an Alpha- Conotoxin Pnia Variant
Nat.Struct.Mol.Biol., 12, 2005
|
|
6S7S
 
 | | Fumarate hydratase of Mycobacterium tuberculosis in complex with formate and allosteric modulator N-(2-Methoxy-5-(N-phenylsulfamoyl)phenyl)-2-(4-oxo-3,4-dihydrophthalazin-1-yl)acetamide | | Descriptor: | FORMIC ACID, Fumarate hydratase class II, ~{N}-[2-methoxy-5-(phenylsulfamoyl)phenyl]-2-(4-oxidanylidene-3~{H}-phthalazin-1-yl)ethanamide | | Authors: | Whitehouse, A.J, Libardo, M.D, Kasbekar, M, Brear, P, Fischer, G, Thomas, C.J, Barry, C.E, Boshoff, H.I, Coyne, A.G, Abell, C. | | Deposit date: | 2019-07-05 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Targeting of Fumarate Hydratase fromMycobacterium tuberculosisUsing Allosteric Inhibitors with a Dimeric-Binding Mode.
J.Med.Chem., 62, 2019
|
|
6S84
 
 | | TsaBDE complex from Thermotoga maritima | | Descriptor: | ATPase YjeE, predicted to have essential role in cell wall biosynthesis, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, ... | | Authors: | Missoury, S, Li-de-La-Sierra-Gallay, I, van Tilbeurgh, H. | | Deposit date: | 2019-07-08 | | Release date: | 2019-07-17 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | The structure of the TsaB/TsaD/TsaE complex reveals an unexpected mechanism for the bacterial t6A tRNA-modification.
Nucleic Acids Res., 46, 2018
|
|
6BR0
 
 | | Solution NMR structure for CcoTx-I | | Descriptor: | Beta-theraphotoxin-Cm1a | | Authors: | Agwa, A.J, Schroeder, C.I. | | Deposit date: | 2017-11-29 | | Release date: | 2018-05-16 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Gating modifier toxins isolated from spider venom: Modulation of voltage-gated sodium channels and the role of lipid membranes.
J. Biol. Chem., 293, 2018
|
|
1IXK
 
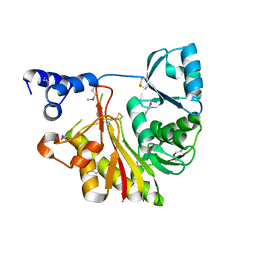 | | Crystal Structure Analysis of Methyltransferase Homolog Protein from Pyrococcus Horikoshii | | Descriptor: | Methyltransferase | | Authors: | Ishikawa, I, Sakai, N, Yao, M, Watanabe, N, Tamura, T, Tanaka, I. | | Deposit date: | 2002-06-25 | | Release date: | 2003-09-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human p120 homologue protein PH1374 from Pyrococcus horikoshii
PROTEINS: STRUCT.,FUNCT.,GENET., 54, 2004
|
|
8UD6
 
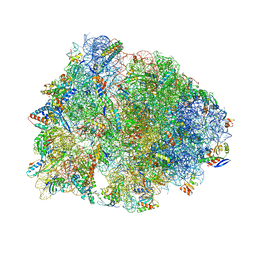 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with cresomycin, mRNA, deacylated A-site tRNAphe, aminoacylated P-site fMet-tRNAmet, and deacylated E-site tRNAphe at 2.70A resolution | | Descriptor: | (4S,5aS,8S,8aR)-4-(2-methylpropyl)-N-[(1R,5Z,7R,8R,9R,10R,11S,12R)-10,11,12-trihydroxy-7-methyl-13-oxa-2-thiabicyclo[7.3.1]tridec-5-en-8-yl]octahydro-2H-oxepino[2,3-c]pyrrole-8-carboxamide (non-preferred name), 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Aleksandrova, E.V, Syroegin, E.A, Wu, K.J.Y, Tresco, B.I.C, Ramkissoon, A, See, D.N.Y, Liow, P, Dittemore, G.A, Yu, M, Testolin, G, Mitcheltree, M.J, Liu, R.Y, Svetlov, M.S, Myers, A.G, Polikanov, Y.S. | | Deposit date: | 2023-09-28 | | Release date: | 2024-02-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An antibiotic preorganized for ribosomal binding overcomes antimicrobial resistance.
Science, 383, 2024
|
|
8U4F
 
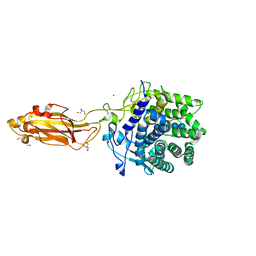 | |
8U49
 
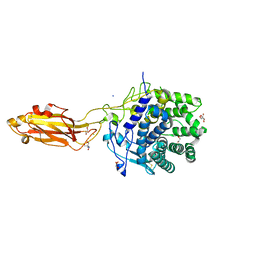 | | The Apo Crystal Structure of BlCel9A from Glycoside Hydrolase Family 9 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Araujo, E.A, Polikarpov, I. | | Deposit date: | 2023-09-10 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular mechanism of cellulose depolymerization by the two-domain BlCel9A enzyme from the glycoside hydrolase family 9.
Carbohydr Polym, 329, 2024
|
|
7S7J
 
 | | Structure of Human SPASTIN-IST1 complex. | | Descriptor: | CALCIUM ION, CHLORIDE ION, IST1 homolog, ... | | Authors: | Skalicky, J.J, Sundquist, W.I. | | Deposit date: | 2021-09-16 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Comprehensive analysis of the human ESCRT-III-MIT domain interactome reveals new cofactors for cytokinetic abscission.
Elife, 11, 2022
|
|
6S88
 
 | | Fumarate hydratase of Mycobacterium tuberculosis in complex with formate and allosteric modulator N-(2-Methoxy-5-((1,2,4,5-tetrahydro-3H-benzo[d]azepin-3-yl)sulfonyl)phenyl)-2-(4-oxo-3,4-dihydrophthalazin-1-yl)acetamide | | Descriptor: | FORMIC ACID, Fumarate hydratase class II, MAGNESIUM ION, ... | | Authors: | Whitehouse, A.J, Libardo, M.D, Kasbekar, M, Brear, P, Fischer, G, Thomas, C.J, Barry, C.E, Boshoff, H.I, Coyne, A.G, Abell, C. | | Deposit date: | 2019-07-08 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Targeting of Fumarate Hydratase fromMycobacterium tuberculosisUsing Allosteric Inhibitors with a Dimeric-Binding Mode.
J.Med.Chem., 62, 2019
|
|
4Z65
 
 | | Carboxypeptidase B with Sulphamoil Arginine | | Descriptor: | Carboxypeptidase B, N~2~-sulfamoyl-L-arginine, ZINC ION | | Authors: | Timofeev, V.I, Akparov, V.K, Kuranova, I.P. | | Deposit date: | 2015-04-03 | | Release date: | 2015-10-14 | | Last modified: | 2015-10-28 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structure of the complex of carboxypeptidase B and N-sulfamoyl-L-arginine.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
1RCO
 
 | | SPINACH RUBISCO IN COMPLEX WITH THE INHIBITOR D-XYLULOSE-2,2-DIOL-1,5-BISPHOSPHATE | | Descriptor: | D-XYLULOSE-2,2-DIOL-1,5-BISPHOSPHATE, RIBULOSE BISPHOSPHATE CARBOXYLASE/OXYGENASE | | Authors: | Taylor, T.C, Andersson, I. | | Deposit date: | 1996-10-31 | | Release date: | 1997-03-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A common structural basis for the inhibition of ribulose 1,5-bisphosphate carboxylase by 4-carboxyarabinitol 1,5-bisphosphate and xylulose 1,5-bisphosphate.
J.Biol.Chem., 271, 1996
|
|
7S8B
 
 | | Cryo-EM structure of human TRPV6 in complex with channel blocker ruthenium red | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, ... | | Authors: | Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2021-09-17 | | Release date: | 2021-11-17 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | Structural mechanisms of TRPV6 inhibition by ruthenium red and econazole.
Nat Commun, 12, 2021
|
|
2XR5
 
 | | Crystal structure of the complex of the carbohydrate recognition domain of human DC-SIGN with pseudo dimannoside mimic. | | Descriptor: | CALCIUM ION, CD209 ANTIGEN, CHLORIDE ION, ... | | Authors: | Thepaut, M, Suitkeviciute, I, Sattin, S, Reina, J, Bernardi, A, Fieschi, F. | | Deposit date: | 2010-09-10 | | Release date: | 2011-10-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structure of a Glycomimetic Ligand in the Carbohydrate Recognition Domain of C-Type Lectin Dc-Sign. Structural Requirements for Selectivity and Ligand Design.
J.Am.Chem.Soc., 135, 2013
|
|
6S42
 
 | | The double mutant(Ile44Leu+Gln102His) of haloalkane dehalogenase DbeA from Bradyrhizobium elkanii USDA94 with an eliminated halide-binding site | | Descriptor: | CHLORIDE ION, HEXANE-1,6-DIOL, Haloalkane dehalogenase, ... | | Authors: | Pudnikova, T, Mesters, J.R, Kuta Smatanova, I. | | Deposit date: | 2019-06-26 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystallization and Crystallographic Analysis of a Bradyrhizobium Elkanii USDA94 Haloalkane Dehalogenase Variant with an Eliminated Halide-Binding Site
Crystals, 2019
|
|
6RV5
 
 | | X-ray structure of the levansucrase from Erwinia tasmaniensis in complex with levanbiose | | Descriptor: | GLYCEROL, Levansucrase (Beta-D-fructofuranosyl transferase), ZINC ION, ... | | Authors: | Polsinelli, I, Caliandro, R, Demitri, N, Benini, S. | | Deposit date: | 2019-05-31 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The Structure of Sucrose-Soaked Levansucrase Crystals fromErwinia tasmaniensisreveals a Binding Pocket for Levanbiose.
Int J Mol Sci, 21, 2019
|
|
4BNE
 
 | | Pacsin2 Interacts with Membranes and Actin-Filaments | | Descriptor: | PROTEIN KINASE C AND CASEIN KINASE SUBSTRATE IN NEURONS PROTEIN 2, SULFATE ION, TRIETHYLENE GLYCOL | | Authors: | Kostan, J, Salzer, U, Orlova, A, Toeroe, I, Hodnik, V, Schreiner, C, Merilainen, J, Nikki, M, Virtanen, I, Lehto, V.-P, Anderluh, G, Egelman, E.H, Djinovic-Carugo, K. | | Deposit date: | 2013-05-15 | | Release date: | 2014-05-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Direct Interaction of Actin Filaments with F-Bar Protein Pacsin2.
Embo Rep., 15, 2014
|
|
6UNG
 
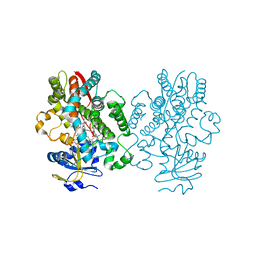 | | Human CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, tert-butyl [(2S)-1-(naphthalen-1-yl)-3-{[(2R)-3-oxo-2-(phenylamino)-3-{[(pyridin-3-yl)methyl]amino}propyl]sulfanyl}propan-2-yl]carbamate | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2019-10-11 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | An increase in side-group hydrophobicity largely improves the potency of ritonavir-like inhibitors of CYP3A4.
Bioorg.Med.Chem., 28, 2020
|
|
2LFN
 
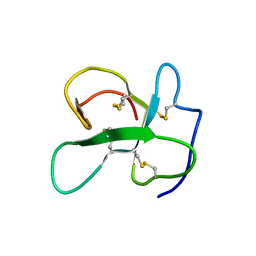 | | Identification of the key regions that drive functional amyloid formation by the fungal hydrophobin EAS | | Descriptor: | Hydrophobin | | Authors: | Macindoe, I, Kwan, A.H, Morris, V.K, Mackay, J.P, Sunde, M. | | Deposit date: | 2011-07-06 | | Release date: | 2012-01-25 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Self-assembly of functional, amphipathic amyloid monolayers by the fungal hydrophobin EAS
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6UNJ
 
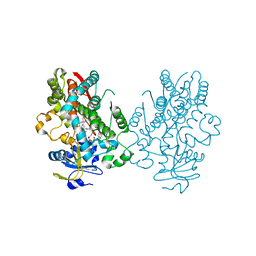 | | Human CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, tert-butyl [(2R)-1-(naphthalen-1-yl)-3-{[(2S)-1-oxo-3-phenyl-1-{[2-(pyridin-3-yl)ethyl]amino}propan-2-yl]sulfanyl}propan-2-yl]carbamate | | Authors: | Sevrioukova, I. | | Deposit date: | 2019-10-12 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An increase in side-group hydrophobicity largely improves the potency of ritonavir-like inhibitors of CYP3A4.
Bioorg.Med.Chem., 28, 2020
|
|
2L0P
 
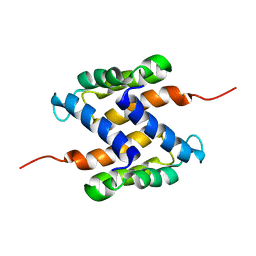 | | Solution structure of human apo-S100A1 protein by NMR spectroscopy | | Descriptor: | S100 calcium binding protein A1 | | Authors: | Nowakowski, M, Jaremko, L, Jaremko, M, Bierzynski, A, Zhukov, I, Ejchart, A. | | Deposit date: | 2010-07-12 | | Release date: | 2011-04-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure and dynamics of human apo-S100A1 protein.
J.Struct.Biol., 174, 2011
|
|
7SJS
 
 | |
1NX8
 
 | | Structure of carbapenem synthase (CarC) complexed with N-acetyl proline | | Descriptor: | 1-ACETYL-L-PROLINE, 2-OXOGLUTARIC ACID, Carbapenem synthase, ... | | Authors: | Clifton, I.J, Doan, L.X, Sleeman, M.C, Topf, M, Suzuki, H, Wilmouth, R.C, Schofield, C.J. | | Deposit date: | 2003-02-10 | | Release date: | 2003-06-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of carbapenem synthase (CarC).
J.Biol.Chem., 278, 2003
|
|
