6UUL
 
 | | Crystal structure of broad and potent HIV-1 neutralizing antibody 438-D5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, D5 Fab Heavy Chain, ... | | Authors: | Kumar, S, Wilson, I.A. | | Deposit date: | 2019-10-30 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | A V H 1-69 antibody lineage from an infected Chinese donor potently neutralizes HIV-1 by targeting the V3 glycan supersite.
Sci Adv, 6, 2020
|
|
6RZ2
 
 | | SalL with Chloroadenosine | | Descriptor: | 5'-CHLORO-5'-DEOXYADENOSINE, Adenosyl-chloride synthase | | Authors: | McKean, I, Frese, A, Cuetos, A, Burley, G, Grogan, G. | | Deposit date: | 2019-06-12 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | S-Adenosyl Methionine Cofactor Modifications Enhance the Biocatalytic Repertoire of Small Molecule C-Alkylation.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
7R33
 
 | | Difference-refined structure of fatty acid photodecarboxylase 20 ps following 400-nm laser irradiation of the dark-state determined by SFX | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid photodecarboxylase, chloroplastic, ... | | Authors: | Hadjidemetriou, K, Coquelle, N, Barends, T.R.M, De Zitter, E, Schlichting, I, Colletier, J.P, Weik, M. | | Deposit date: | 2022-02-06 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Time-resolved serial femtosecond crystallography on fatty-acid photodecarboxylase: lessons learned.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7R34
 
 | | Difference-refined structure of fatty acid photodecarboxylase 900 ps following 400-nm laser irradiation of the dark-state determined by SFX | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid photodecarboxylase, chloroplastic, ... | | Authors: | Hadjidemetriou, K, Coquelle, N, Barends, T.R.M, De Zitter, E, Schlichting, I, Colletier, J.P, Weik, M. | | Deposit date: | 2022-02-06 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Time-resolved serial femtosecond crystallography on fatty-acid photodecarboxylase: lessons learned.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7R35
 
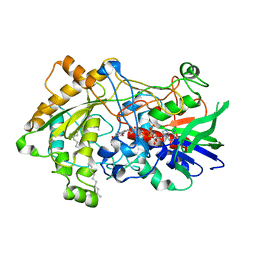 | | Difference-refined structure of fatty acid photodecarboxylase 300 ns following 400-nm laser irradiation of the dark-state determined by SFX | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid photodecarboxylase, chloroplastic, ... | | Authors: | Hadjidemetriou, K, Coquelle, N, Barends, T.R.M, De Zitter, E, Schlichting, I, Colletier, J.P, Weik, W. | | Deposit date: | 2022-02-06 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Time-resolved serial femtosecond crystallography on fatty-acid photodecarboxylase: lessons learned.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
2XRI
 
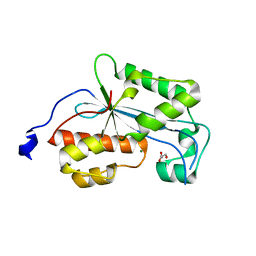 | | Crystal structure of human ERI1 exoribonuclease 3 | | Descriptor: | ERI1 EXORIBONUCLEASE 3, GLYCEROL, MAGNESIUM ION | | Authors: | Welin, M, Siponen, M.I, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Karlberg, T, Kol, S, Kotenyova, T, Kouznetsova, E, Moche, M, Nyman, T, Persson, C, Schuler, H, Schutz, P, Thorsell, A.G, Tresaugues, L, Van Der Berg, S, Wahlberg, E, Weigelt, J, Nordlund, P. | | Deposit date: | 2010-09-15 | | Release date: | 2010-09-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of Human Eri1 Exoribonuclease 3
To be Published
|
|
3PGM
 
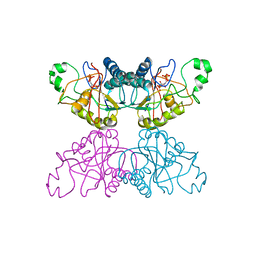 | | THE STRUCTURE OF YEAST PHOSPHOGLYCERATE MUTASE AT 0.28 NM RESOLUTION | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, Phosphoglycerate mutase 1, SULFATE ION | | Authors: | Campbell, J.W, Hodgson, G.I, Warwicker, J, Winn, S.I, Watson, H.C. | | Deposit date: | 1982-04-06 | | Release date: | 1982-05-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and activity of phosphoglycerate mutase.
Philos.Trans.R.Soc.London,Ser.B, 293, 1981
|
|
6S1S
 
 | | Crystal structure of AmpC from Pseudomonas aeruginosa in complex with [3-(2-carboxyvinyl)phenyl]boronic acid] inhibitor | | Descriptor: | (~{E})-3-[3-(dihydroxyboranyl)phenyl]prop-2-enoic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Kekez, I, Vicario, M, Bellio, P, Tosoni, E, Celenza, G, Blazquez, J, Tondi, D, Cendron, L. | | Deposit date: | 2019-06-19 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Phenylboronic Acids Probing Molecular Recognition against Class A and Class C beta-lactamases.
Antibiotics, 8, 2019
|
|
6UYD
 
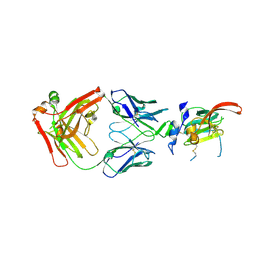 | | Structure of Hepatitis C Virus Envelope Glycoprotein E2mc3-v1 redesigned core from genotype 1a bound to broadly neutralizing antibody AR3C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein E2, ... | | Authors: | Tzarum, N, Wilson, I.A, Zhu, J. | | Deposit date: | 2019-11-13 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | Proof of concept for rational design of hepatitis C virus E2 core nanoparticle vaccines.
Sci Adv, 6, 2020
|
|
2JIP
 
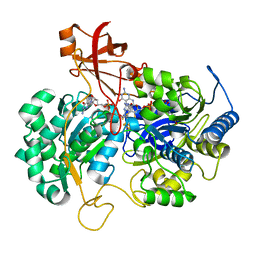 | | A New Catalytic Mechanism of Periplasmic Nitrate Reductase from Desulfovibrio desulfuricans ATCC 27774 from Crystallographic and EPR Data and based on detailed analysis of the sixth ligand | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, IRON/SULFUR CLUSTER, MOLYBDENUM ATOM, ... | | Authors: | Najmudin, S, Gonzalez, P.J, Trincao, J, Coelho, C, Mukhopadhyay, A, Romao, C.C, Moura, I, Moura, J.J.G, Brondino, C.D, Romao, M.J. | | Deposit date: | 2007-06-28 | | Release date: | 2008-03-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Periplasmic Nitrate Reductase Revisited: A Sulfur Atom Completes the Sixth Coordination of the Catalytic Molybdenum.
J.Biol.Inorg.Chem., 13, 2008
|
|
5OBX
 
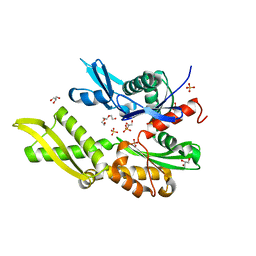 | | Mycoplasma genitalium DnaK-NBD | | Descriptor: | Chaperone protein DnaK, GLYCEROL, SULFATE ION, ... | | Authors: | Adell, M, Calisto, B, Fita, I, Martinelli, L. | | Deposit date: | 2017-06-29 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The nucleotide-bound/substrate-bound conformation of the Mycoplasma genitalium DnaK chaperone.
Protein Sci., 27, 2018
|
|
2JDT
 
 | | Structure of PKA-PKB chimera complexed with ISOQUINOLINE-5-SULFONIC ACID (2-(2-(4-CHLOROBENZYLOXY) ETHYLAMINO)ETHYL)AMIDE | | Descriptor: | CAMP-DEPENDENT PROTEIN KINASE, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA, ISOQUINOLINE-5-SULFONIC ACID (2-(2-(4-CHLOROBENZYLOXY)ETHYLAMINO)ETHYL)AMIDE | | Authors: | Davies, T.G, Verdonk, M.L, Graham, B, Saalau-Bethell, S, Hamlett, C.C.F, McHardy, T, Collins, I, Garrett, M.D, Workman, P, Woodhead, S.J, Jhoti, H, Barford, D. | | Deposit date: | 2007-01-12 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A Structural Comparison of Inhibitor Binding to Pkb, Pka and Pka-Pkb Chimera
J.Mol.Biol., 367, 2007
|
|
6KZ0
 
 | | HRV14 3C in complex with single chain antibody GGVV | | Descriptor: | GGVV H chain, GGVV L chain, Genome polyprotein | | Authors: | Meng, B, Yang, B, Wilson, I.A. | | Deposit date: | 2019-09-21 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibitory antibodies identify unique sites of therapeutic vulnerability in rhinovirus and other enteroviruses.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1NKW
 
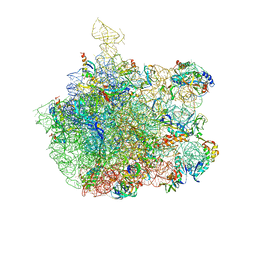 | | Crystal Structure Of The Large Ribosomal Subunit From Deinococcus Radiodurans | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L11, 50S ribosomal protein L13, ... | | Authors: | Harms, J.M, Schluenzen, F, Zarivach, R, Bashan, A, Gat, S, Agmon, I, Bartels, H, Franceschi, F, Yonath, A. | | Deposit date: | 2003-01-05 | | Release date: | 2003-02-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | High resolution structure of the large ribosomal subunit from a mesophilic eubacterium
Cell(Cambridge,Mass.), 107, 2001
|
|
2JIO
 
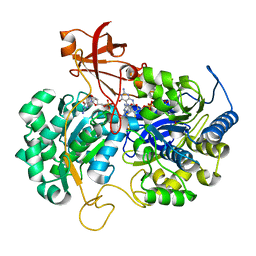 | | A New Catalytic Mechanism of Periplasmic Nitrate Reductase from Desulfovibrio desulfuricans ATCC 27774 from Crystallographic and EPR Data and based on detailed analysis of the sixth ligand | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, IRON/SULFUR CLUSTER, MOLYBDENUM ATOM, ... | | Authors: | Najmudin, S, Gonzalez, P.J, Trincao, J, Coelho, C, Mukhopadhyay, A, Romao, C.C, Moura, I, Moura, J.J.G, Brondino, C.D, Romao, M.J. | | Deposit date: | 2007-06-28 | | Release date: | 2008-03-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Periplasmic Nitrate Reductase Revisited: A Sulfur Atom Completes the Sixth Coordination of the Catalytic Molybdenum.
J.Biol.Inorg.Chem., 13, 2008
|
|
5OH4
 
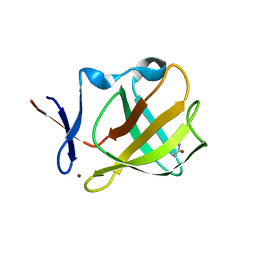 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with Piperidine-2,6-dione (Glutarimide) | | Descriptor: | Cereblon isoform 4, ZINC ION, piperidine-2,6-dione | | Authors: | Boichenko, I, Albrecht, R, Lupas, A.N, Hernandez Alvarez, B, Hartmann, M.D. | | Deposit date: | 2017-07-14 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Chemical Ligand Space of Cereblon.
Acs Omega, 3, 2018
|
|
7RKL
 
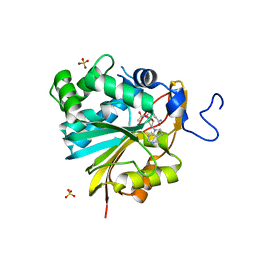 | | Structure of Nicotinamide N-Methyltransferase (NNMT) in complex with II399 (P1 space group) | | Descriptor: | 3-[3-(acetyl{[(1R,2R,3S,4R)-4-(4-chloro-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-2,3-dihydroxycyclopentyl]methyl}amino)prop-1-yn-1-yl]benzamide, NNMT protein, SULFATE ION | | Authors: | Yadav, R, Noinaj, N, Iyamu, I.D, Huang, R. | | Deposit date: | 2021-07-22 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Exploring Unconventional SAM Analogues To Build Cell-Potent Bisubstrate Inhibitors for Nicotinamide N-Methyltransferase.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
6S7Z
 
 | | Fumarate hydratase of Mycobacterium tuberculosis in complex with formate and allosteric modulator N-(5-((3,4-Dihydroisoquinolin-2(1H)-yl)sulfonyl)-2-methoxyphenyl)-2-(4-oxo-3,4-dihydrophthalazin-1-yl)acetamide | | Descriptor: | FORMIC ACID, Fumarate hydratase class II, MAGNESIUM ION, ... | | Authors: | Whitehouse, A.J, Libardo, M.D, Kasbekar, M, Brear, P, Fischer, G, Thomas, C.J, Barry, C.E, Boshoff, H.I, Coyne, A.G, Abell, C. | | Deposit date: | 2019-07-07 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Targeting of Fumarate Hydratase fromMycobacterium tuberculosisUsing Allosteric Inhibitors with a Dimeric-Binding Mode.
J.Med.Chem., 62, 2019
|
|
5CN4
 
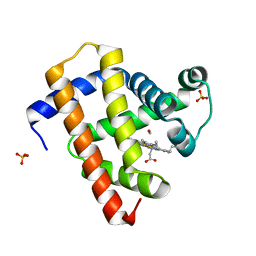 | | Ultrafast dynamics in myoglobin: -0.1 ps time delay | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Barends, T.R.M, Foucar, L, Ardevol, A, Nass, K.J, Aquila, A, Botha, S, Doak, R.B, Falahati, K, Hartmann, E, Hilpert, M, Heinz, M, Hoffmann, M.C, Koefinger, J, Koglin, J, Kovacsova, G, Liang, M, Milathianaki, D, Lemke, H.T, Reinstein, J, Roome, C.M, Shoeman, R.L, Williams, G.J, Burghardt, I, Hummer, G, Boutet, S, Schlichting, I. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Direct observation of ultrafast collective motions in CO myoglobin upon ligand dissociation.
Science, 350, 2015
|
|
7K76
 
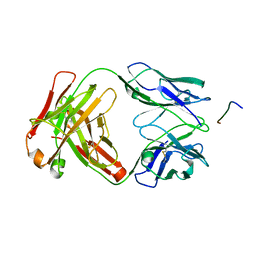 | |
1NDA
 
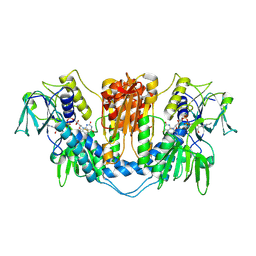 | | THE STRUCTURE OF TRYPANOSOMA CRUZI TRYPANOTHIONE REDUCTASE IN THE OXIDIZED AND NADPH REDUCED STATE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, TRYPANOTHIONE OXIDOREDUCTASE | | Authors: | Lantwin, C.B, Kabsch, W, Pai, E.F, Schlichting, I, Krauth-Siegel, R.L. | | Deposit date: | 1993-07-02 | | Release date: | 1994-09-30 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The structure of Trypanosoma cruzi trypanothione reductase in the oxidized and NADPH reduced state.
Proteins, 18, 1994
|
|
2JGT
 
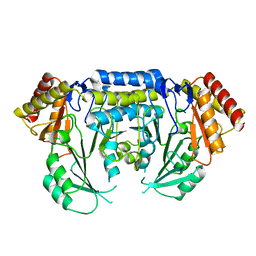 | | Low resolution structure of SPT | | Descriptor: | SERINE PALMITOYLTRANSFERASE | | Authors: | Yard, B.A, Carter, L.G, Johnson, K.A, Overton, I.M, Mcmahon, S.A, Dorward, M, Liu, H, Puech, D, Oke, M, Barton, G.J, Naismith, J.H, Campopiano, D.J. | | Deposit date: | 2007-02-14 | | Release date: | 2007-05-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Structure of Serine Palmitoyltransferase; Gateway to Sphingolipid Biosynthesis.
J.Mol.Biol., 370, 2007
|
|
8TR5
 
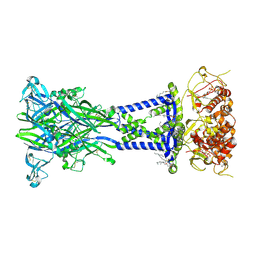 | | Cryo-EM structure of the rat P2X7 receptor in the apo closed state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GUANOSINE-5'-DIPHOSPHATE, P2X purinoceptor 7, ... | | Authors: | Oken, A.C, Lisi, N.E, Krishnamurthy, I, McCarthy, A.E, Godsey, M.H, Glasfeld, A, Mansoor, S.E. | | Deposit date: | 2023-08-09 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | High-affinity agonism at the P2X 7 receptor is mediated by three residues outside the orthosteric pocket.
Nat Commun, 15, 2024
|
|
8TRJ
 
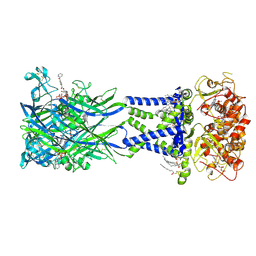 | | Cryo-EM structure of the rat P2X7 receptor in complex with the high-affinity agonist BzATP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3'-O-(4-benzoylbenzoyl)adenosine 5'-(tetrahydrogen triphosphate), GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Oken, A.C, Lisi, N.E, Krishnamurthy, I, McCarthy, A.E, Godsey, M.H, Glasfeld, A, Mansoor, S.E. | | Deposit date: | 2023-08-09 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | High-affinity agonism at the P2X 7 receptor is mediated by three residues outside the orthosteric pocket.
Nat Commun, 15, 2024
|
|
6RSM
 
 | | Solution NMR structure of the peptide 12530 from medicinal leech Hirudo medicinalis in dodecylphosphocholine micelles | | Descriptor: | peptide 12530 | | Authors: | Talyzina, I.A, Nadezhdin, K.D, Grafskaia, E.N, Arseniev, A.S, Lazarev, V.N. | | Deposit date: | 2019-05-21 | | Release date: | 2019-07-24 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Medicinal leech antimicrobial peptides lacking toxicity represent a promising alternative strategy to combat antibiotic-resistant pathogens.
Eur.J.Med.Chem., 180, 2019
|
|
