4J14
 
 | | Crystal Structure of Human Cytochrome P450 CYP46A1 with Posaconazole Bound | | Descriptor: | Cholesterol 24-hydroxylase, GLYCEROL, POSACONAZOLE, ... | | Authors: | Stout, C.D, Mast, N, Pikuleva, I.A. | | Deposit date: | 2013-01-31 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Antifungal Azoles: Structural Insights into Undesired Tight Binding to Cholesterol-Metabolizing CYP46A1.
Mol.Pharmacol., 84, 2013
|
|
4J2O
 
 | | Crystal structure of NADP-bound WbjB from A. baumannii community strain D1279779 | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, UDP-N-acetylglucosamine 4,6-dehydratase/5-epimerase | | Authors: | Shah, B.S, Harrop, S.J, Paulsen, I.T, Mabbutt, B.C. | | Deposit date: | 2013-02-05 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.653 Å) | | Cite: | Crystal structure of a UDP-GlcNAc epimerase for surface polysaccharide biosynthesis in Acinetobacter baumannii.
Plos One, 13, 2018
|
|
1QRP
 
 | | Human pepsin 3A in complex with a phosphonate inhibitor IVA-VAL-VAL-LEU(P)-(O)PHE-ALA-ALA-OME | | Descriptor: | PEPSIN 3A, methyl N-[(2S)-2-({(S)-hydroxy[(1R)-3-methyl-1-{[N-(3-methylbutanoyl)-L-valyl-L-valyl]amino}butyl]phosphoryl}oxy)-3-phenylpropanoyl]-L-alanyl-L-alaninate | | Authors: | Fujinaga, M, Cherney, M.M, Tarasova, N.I, Bartlett, P.A, Hanson, J.E, James, M.N.G. | | Deposit date: | 1999-06-15 | | Release date: | 1999-06-18 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural study of the complex between human pepsin and a phosphorus-containing peptidic -transition-state analog.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
3G9X
 
 | | Structure of haloalkane dehalogenase DhaA14 mutant I135F from Rhodococcus rhodochrous | | Descriptor: | ACETATE ION, CHLORIDE ION, Haloalkane dehalogenase, ... | | Authors: | Gavira, J.A, Stsiapanava, A, Kuty, M, Lapkouski, M, Dohnalek, J, Kuta Smatanova, I. | | Deposit date: | 2009-02-15 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Atomic resolution studies of haloalkane dehalogenases DhaA04, DhaA14 and DhaA15 with engineered access tunnels.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
8ALS
 
 | |
3GEW
 
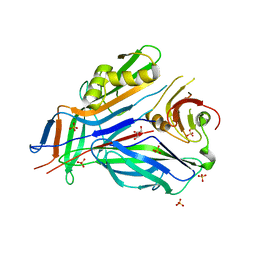 | | FaeE-FaeG chaperone-major pilin complex of F4 ad fimbriae | | Descriptor: | Chaperone protein faeE, GLYCEROL, K88 fimbrial protein AD, ... | | Authors: | Van Molle, I, Moonens, K, Garcia-Pino, A, Buts, L, Bouckaert, J, De Greve, H. | | Deposit date: | 2009-02-26 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and thermodynamic characterization of pre- and postpolymerization states in the F4 fimbrial subunit FaeG
J.Mol.Biol., 394, 2009
|
|
3P4Z
 
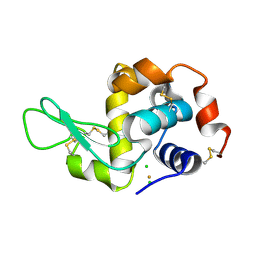 | | Time-dependent and Protein-directed In Situ Growth of Gold Nanoparticles in a Single Crystal of Lysozyme | | Descriptor: | CHLORIDE ION, GOLD 3+ ION, GOLD ION, ... | | Authors: | Wei, H, Wang, Z, Zhang, J, House, S, Gao, Y.-G, Yang, L, Robinson, H, Tan, L.H, Xing, H, Hou, C, Robertson, I.M, Zuo, J.-M, Lu, Y. | | Deposit date: | 2010-10-07 | | Release date: | 2011-02-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Time-dependent, protein-directed growth of gold nanoparticles within a single crystal of lysozyme.
Nat Nanotechnol, 6, 2011
|
|
5W0J
 
 | |
5KK2
 
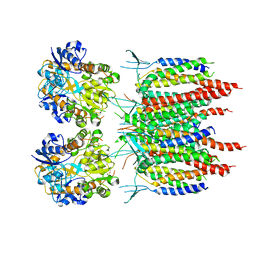 | | Architecture of fully occupied GluA2 AMPA receptor - TARP complex elucidated by single particle cryo-electron microscopy | | Descriptor: | Glutamate receptor 2, Voltage-dependent calcium channel gamma-2 subunit | | Authors: | Zhao, Y, Chen, S, Yoshioka, C, Baconguis, I, Gouaux, E. | | Deposit date: | 2016-06-20 | | Release date: | 2016-07-06 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Architecture of fully occupied GluA2 AMPA receptor-TARP complex elucidated by cryo-EM.
Nature, 536, 2016
|
|
7LZH
 
 | | Structure of the glutamate receptor-like channel AtGLR3.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Gangwar, S.P, Green, M.N, Sobolevsky, A.I. | | Deposit date: | 2021-03-09 | | Release date: | 2021-07-28 | | Last modified: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Structure of the Arabidopsis thaliana glutamate receptor-like channel GLR3.4.
Mol.Cell, 81, 2021
|
|
6GN5
 
 | | CRYSTAL STRUCTURE OF HUMAN GRAMD1C START DOMAIN | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GRAM domain-containing protein 1C | | Authors: | Friese, A, Vetter, I.R. | | Deposit date: | 2018-05-30 | | Release date: | 2019-06-19 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | The cholesterol transfer protein GRAMD1A regulates autophagosome biogenesis.
Nat.Chem.Biol., 15, 2019
|
|
8AIE
 
 | | Crystal structure of D-amino acid aminotransferase from Aminobacterium colombiense complexed with D-cycloserine | | Descriptor: | 3-azanyloxy-2-[(~{E})-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]propanoic acid, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Aminotransferase class IV, ... | | Authors: | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Shilova, S.A, Popov, V.O, Bezsudnova, E.Y. | | Deposit date: | 2022-07-26 | | Release date: | 2022-11-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 3D Structure of D-Аmino Acid Тransaminase from Aminobacterium colombiense in Complex with D-Cycloserine
Crystallography Reports, 68, 2023
|
|
8AHR
 
 | | Crystal structure of D-amino acid aminotransferase from Aminobacterium colombiense in holo form with PLP | | Descriptor: | Aminotransferase class IV, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Shilova, S.A, Rakitina, T.V, Popov, V.O, Bezsudnova, E.Y. | | Deposit date: | 2022-07-22 | | Release date: | 2022-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | To the Understanding of Catalysis by D-Amino Acid Transaminases: A Case Study of the Enzyme from Aminobacterium colombiense.
Molecules, 28, 2023
|
|
3HJB
 
 | | 1.5 Angstrom Crystal Structure of Glucose-6-phosphate Isomerase from Vibrio cholerae. | | Descriptor: | CALCIUM ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-05-21 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.5 Angstrom Crystal Structure of Glucose-6-phosphate Isomerase from Vibrio cholerae.
To be Published
|
|
1QQ3
 
 | | THE SOLUTION STRUCTURE OF THE HEME BINDING VARIANT ARG98CYS OF OXIDIZED ESCHERICHIA COLI CYTOCHROME B562 | | Descriptor: | CYTOCHROME B562, HEME B/C | | Authors: | Arnesano, F, Banci, L, Bertini, I, Ciofi-Baffoni, S, Barker, P.D, Woodyear, T. | | Deposit date: | 1999-06-10 | | Release date: | 2000-05-24 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Structural consequences of b- to c-type heme conversion in oxidized Escherichia coli cytochrome b562.
Biochemistry, 39, 2000
|
|
3OUM
 
 | | Crystal Structure of toxoflavin-degrading enzyme in complex with toxoflavin | | Descriptor: | 1,6-dimethylpyrimido[5,4-e][1,2,4]triazine-5,7(1H,6H)-dione, MANGANESE (II) ION, toxoflavin-degrading enzyme | | Authors: | Kim, M.I, Rhee, S. | | Deposit date: | 2010-09-15 | | Release date: | 2011-08-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional analysis of phytotoxin toxoflavin-degrading enzyme
Plos One, 6, 2011
|
|
6GSP
 
 | | RIBONUCLEASE T1/3'-GMP, 15 WEEKS | | Descriptor: | CALCIUM ION, GUANOSINE-3'-MONOPHOSPHATE, RIBONUCLEASE T1 | | Authors: | Zegers, I, Wyns, L. | | Deposit date: | 1997-12-09 | | Release date: | 1998-03-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Hydrolysis of a slow cyclic thiophosphate substrate of RNase T1 analyzed by time-resolved crystallography.
Nat.Struct.Biol., 5, 1998
|
|
4G6A
 
 | |
6H5Z
 
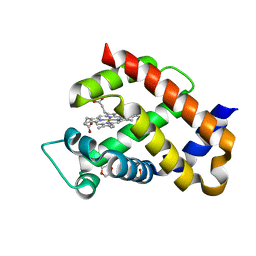 | | Ferric murine neuroglobin F106A mutant | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Neuroglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Exertier, C, Vallone, B, Savino, C, Freda, I, Montemiglio, L.C, Cerutti, G, Scaglione, A, Parisi, G. | | Deposit date: | 2018-07-25 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Proximal and distal control for ligand binding in neuroglobin: role of the CD loop and evidence for His64 gating.
Sci Rep, 9, 2019
|
|
3OUL
 
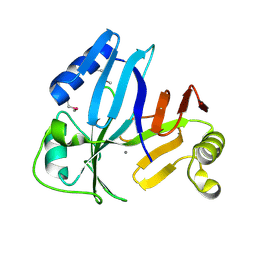 | |
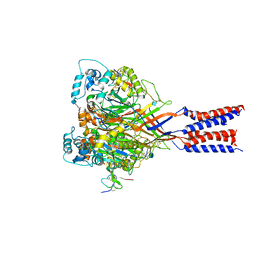
4FZ0
 
 | |
8AYK
 
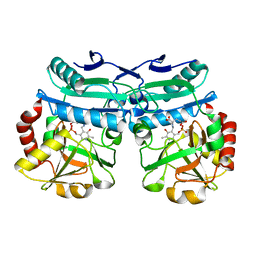 | | Crystal structure of D-amino acid aminotrensferase from Aminobacterium colombiense complexed with D-glutamate | | Descriptor: | (~{Z})-2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]pent-2-enedioic acid, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Aminotransferase class IV | | Authors: | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Shilova, S.A, Rakitina, T.V, Minyaev, M.E, Popov, V.O, Bezsudnova, E.Y. | | Deposit date: | 2022-09-02 | | Release date: | 2022-11-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | To the Understanding of Catalysis by D-Amino Acid Transaminases: A Case Study of the Enzyme from Aminobacterium colombiense.
Molecules, 28, 2023
|
|
4JPJ
 
 | | Crystal structure of the germline-targeting HIV-1 gp120 engineered outer domain, eOD-GT6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Germline-targeting HIV-1 gp120 engineered outer domain, eOD-GT6 | | Authors: | Julien, J.-P, Jardine, J, Schief, W.R, Wilson, I.A. | | Deposit date: | 2013-03-19 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Rational HIV immunogen design to target specific germline B cell receptors.
Science, 340, 2013
|
|
2XIG
 
 | | The structure of the Helicobacter pylori ferric uptake regulator Fur reveals three functional metal binding sites | | Descriptor: | CITRIC ACID, FERRIC UPTAKE REGULATION PROTEIN, ZINC ION | | Authors: | Dian, C, Vitale, S, Leonard, G.A, Fauquant, F, Muller, C, Bahlawane, C, de Reuse, H, Michaud-Soret, I, Terradot, L. | | Deposit date: | 2010-06-29 | | Release date: | 2011-01-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Structure of the Helicobacter Pylori Ferric Uptake Regulator Fur Reveals Three Functional Metal Binding Sites.
Mol.Microbiol., 79, 2011
|
|
2Y0D
 
 | | BceC mutation Y10K | | Descriptor: | SULFATE ION, UDP-GLUCOSE DEHYDROGENASE, URIDINE-5'-DIPHOSPHATE-GLUCURONIC ACID | | Authors: | Rocha, J, Popescu, A.O, Borges, P, Mil-Homens, D, Sa-Correia, I, Fialho, A.M, Frazao, C. | | Deposit date: | 2010-12-02 | | Release date: | 2011-07-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Burkholderia Cepacia Udp-Glucose Dehydrogenase (Ugd) Bcec and Role of Tyr10 in Final Hydrolysis of Ugd Thioester Intermediate.
J.Bacteriol., 193, 2011
|
|
