8SMN
 
 | | Xenopus laevis hyaluronan synthase 1, nascent HA polymer bound state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab15 heavy chain, Fab15 light chain, ... | | Authors: | Gorniak, I, Zimmer, J. | | Deposit date: | 2023-04-26 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Atomistic insights into hyaluronan synthesis, secretion, and length control
To Be Published
|
|
8SMM
 
 | | Xenopus laevis hyaluronan synthase 1 | | Descriptor: | Fab15 heavy chain, Fab15 light chain, Hyaluronan synthase 1 | | Authors: | Gorniak, I, Zimmer, J. | | Deposit date: | 2023-04-26 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Atomistic insights into hyaluronan synthesis, secretion, and length control
To Be Published
|
|
8SMP
 
 | | Xenopus laevis hyaluronan synthase 1, UDP-bound, gating loop inserted state | | Descriptor: | Fab15 heavy chain, Fab15 light chain, Hyaluronan synthase 1, ... | | Authors: | Gorniak, I, Zimmer, J. | | Deposit date: | 2023-04-26 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Atomistic insights into hyaluronan synthesis, secretion, and length control
To Be Published
|
|
1MH5
 
 | | The Structure Of The Complex Of The Fab Fragment Of The Esterolytic Antibody MS6-164 and A Transition-State Analog | | Descriptor: | IMMUNOGLOBULIN MS6-164, N-{[2-({[1-(4-CARBOXYBUTANOYL)AMINO]-2-PHENYLETHYL}-HYDROXYPHOSPHINYL)OXY]ACETYL}-2-PHENYLETHYLAMINE, SULFATE ION | | Authors: | Ruzheinikov, S.N, Muranova, T.A, Sedelnikova, S.E, Partridge, L.J, Blackburn, G.M, Murray, I.A, Kakinuma, H, Takashi, N, Shimazaki, K, Sun, J, Nishi, Y, Rice, D.W. | | Deposit date: | 2002-08-19 | | Release date: | 2003-09-23 | | Last modified: | 2011-11-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-resolution crystal structure of the Fab-fragments of a family of mouse catalytic antibodies with esterase activity
J.Mol.Biol., 332, 2003
|
|
1M9X
 
 | | X-ray crystal structure of Cyclophilin A/HIV-1 CA N-terminal domain (1-146) M-type H87A,A88M,G89A Complex. | | Descriptor: | Cyclophilin A, HIV-1 Capsid | | Authors: | Howard, B.R, Vajdos, F.F, Li, S, Sundquist, W.I, Hill, C.P. | | Deposit date: | 2002-07-30 | | Release date: | 2003-05-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the catalytic mechanism of cyclophilin A
Nat.Struct.Biol., 10, 2003
|
|
7U5G
 
 | | ACS122 Fab | | Descriptor: | ACS122 Fab Heavy chain, ACS122 Fab Light chain | | Authors: | Farokhi, E, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2022-03-02 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Complementary antibody lineages achieve neutralization breadth in an HIV-1 infected elite neutralizer.
Plos Pathog., 18, 2022
|
|
1K30
 
 | | Crystal Structure Analysis of Squash (Cucurbita moschata) glycerol-3-phosphate (1)-acyltransferase | | Descriptor: | glycerol-3-phosphate acyltransferase | | Authors: | Turnbull, A.P, Rafferty, J.B, Sedelnikova, S.E, Slabas, A.R, Schierer, T.P, Kroon, J.T, Simon, J.W, Fawcett, T, Nishida, I, Murata, N, Rice, D.W. | | Deposit date: | 2001-10-01 | | Release date: | 2001-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Analysis of the structure, substrate specificity, and mechanism of squash glycerol-3-phosphate (1)-acyltransferase.
Structure, 9, 2001
|
|
1JGN
 
 | | Solution structure of the C-terminal PABC domain of human poly(A)-binding protein in complex with the peptide from Paip2 | | Descriptor: | polyadenylate-binding protein 1, polyadenylate-binding protein-interacting protein 2 | | Authors: | Kozlov, G, Siddiqui, N, Coillet-Matillon, S, Ekiel, I, Gehring, K. | | Deposit date: | 2001-06-26 | | Release date: | 2003-06-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis of ligand recognition by PABC, a highly specific peptide-binding domain found in poly(A)-binding protein and a HECT ubiquitin ligase
EMBO J., 23, 2004
|
|
4N5K
 
 | |
1M9N
 
 | | CRYSTAL STRUCTURE OF THE HOMODIMERIC BIFUNCTIONAL TRANSFORMYLASE AND CYCLOHYDROLASE ENZYME AVIAN ATIC IN COMPLEX WITH AICAR AND XMP AT 1.93 ANGSTROMS. | | Descriptor: | AICAR TRANSFORMYLASE-IMP CYCLOHYDROLASE, AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, POTASSIUM ION, ... | | Authors: | Wolan, D.W, Greasly, S.E, Beardsley, G.P, Wilson, I.A. | | Deposit date: | 2002-07-29 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural Insights into the Avian AICAR Transformylase Mechanism.
Biochemistry, 41, 2002
|
|
1MEJ
 
 | | Human Glycinamide Ribonucleotide Transformylase domain at pH 8.5 | | Descriptor: | GLYCEROL, PHOSPHATE ION, Phosphoribosylglycinamide formyltransferase | | Authors: | Zhang, Y, Desharnais, J, Greasley, S.E, Beardsley, G.P, Boger, D.L, Wilson, I.A. | | Deposit date: | 2002-08-08 | | Release date: | 2002-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of human GAR Tfase of low and high pH and with substrate beta-GAR
Biochemistry, 41, 2002
|
|
4N5J
 
 | |
1MJU
 
 | | 1.22 ANGSTROM RESOLUTION CRYSTAL STRUCTURE OF THE FAB FRAGMENT OF ESTEROLYTIC ANTIBODY MS6-12 | | Descriptor: | GLYCEROL, IMMUNOGLOBULIN MS6-12 | | Authors: | Ruzheinikov, S.N, Muranova, T.A, Sedelnikova, S.E, Partridge, L.J, Blackburn, G.M, Murray, I.A, Kakinuma, H, Takashi, N, Shimazaki, K, Sun, J, Nishi, Y, Rice, D.W. | | Deposit date: | 2002-08-28 | | Release date: | 2003-09-23 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | High-resolution crystal structure of the Fab-fragments of a family of mouse catalytic antibodies with esterase activity
J.Mol.Biol., 332, 2003
|
|
1MKI
 
 | | Crystal Structure of Bacillus Subtilis Probable Glutaminase, APC1040 | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Probable Glutaminase ybgJ | | Authors: | Kim, Y, Dementieva, I, Vinokour, E, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-08-29 | | Release date: | 2003-06-03 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional and structural characterization of four glutaminases from Escherichia coli and Bacillus subtilis.
Biochemistry, 47, 2008
|
|
1MK6
 
 | | SOLUTION STRUCTURE OF THE 8,9-DIHYDRO-8-(N7-GUANYL)-9-HYDROXY-AFLATOXIN B1 ADDUCT MISPAIRED WITH DEOXYADENOSINE | | Descriptor: | 5'-D(*AP*CP*AP*TP*CP*GP*AP*TP*CP*T)-3', 5'-D(*AP*GP*AP*TP*AP*GP*AP*TP*GP*T)-3', 8,9-DIHYDRO-9-HYDROXY-AFLATOXIN B1 | | Authors: | Giri, I, Johnston, D.S, Stone, M.P. | | Deposit date: | 2002-08-28 | | Release date: | 2002-10-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | MISPAIRING OF THE 8,9-DIHYDRO-8-(N7-GUANYL)-9-HYDROXY-AFLATOXIN B1 ADDUCT WITH DEOXYADENOSINE RESULTS IN EXTRUSION OF THE MISMATCHED DA TOWARD THE MAJOR GROOVE
Biochemistry, 41, 2002
|
|
4AY1
 
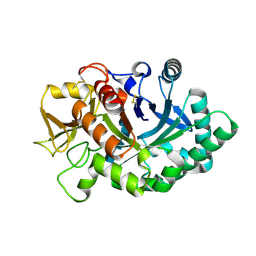 | | Human YKL-39 is a pseudo-chitinase with retained chitooligosaccharide binding properties | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITINASE-3-LIKE PROTEIN 2 | | Authors: | Schimpl, M, Rush, C.L, Betou, M, Eggleston, I.M, Penman, G.A, Recklies, A.D, van Aalten, D.M.F. | | Deposit date: | 2012-06-17 | | Release date: | 2012-08-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Human Ykl-39 is a Pseudo-Chitinase with Retained Chitooligosaccharide-Binding Properties.
Biochem.J., 446, 2012
|
|
1MMN
 
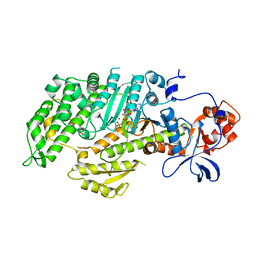 | | X-RAY STRUCTURES OF THE MGADP, MGATPGAMMAS, AND MGAMPPNP COMPLEXES OF THE DICTYOSTELIUM DISCOIDEUM MYOSIN MOTOR DOMAIN | | Descriptor: | MAGNESIUM ION, MYOSIN, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Gulick, A.M, Bauer, C.B, Thoden, J.B, Rayment, I. | | Deposit date: | 1997-07-18 | | Release date: | 1997-12-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray structures of the MgADP, MgATPgammaS, and MgAMPPNP complexes of the Dictyostelium discoideum myosin motor domain.
Biochemistry, 36, 1997
|
|
1MPY
 
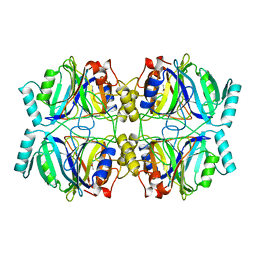 | | STRUCTURE OF CATECHOL 2,3-DIOXYGENASE (METAPYROCATECHASE) FROM PSEUDOMONAS PUTIDA MT-2 | | Descriptor: | ACETONE, CATECHOL 2,3-DIOXYGENASE, FE (II) ION | | Authors: | Kita, A, Kita, S, Fujisawa, I, Inaka, K, Ishida, T, Horiike, K, Nozaki, M, Miki, K. | | Deposit date: | 1998-10-20 | | Release date: | 1999-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | An archetypical extradiol-cleaving catecholic dioxygenase: the crystal structure of catechol 2,3-dioxygenase (metapyrocatechase) from Ppseudomonas putida mt-2.
Structure Fold.Des., 7, 1999
|
|
1MEP
 
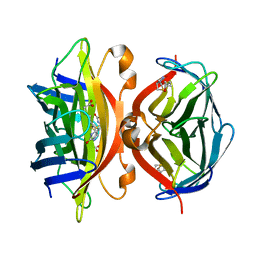 | | Crystal Structure of Streptavidin Double Mutant S45A/D128A with Biotin: Cooperative Hydrogen-Bond Interactions in the Streptavidin-Biotin System. | | Descriptor: | BIOTIN, Streptavidin | | Authors: | Hyre, D.E, Le Trong, I, Merritt, E.A, Stenkamp, R.E, Green, N.M, Stayton, P.S. | | Deposit date: | 2002-08-08 | | Release date: | 2003-09-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Cooperative hydrogen bond interactions in the streptavidin-biotin system
Protein Sci., 15, 2006
|
|
4CXF
 
 | | Structure of CnrH in complex with the cytosolic domain of CnrY | | Descriptor: | CHLORIDE ION, CNRY, RNA POLYMERASE SIGMA FACTOR CNRH, ... | | Authors: | Maillard, A.P, Girard, E, Ziani, W, Petit-Hartlein, I, Kahn, R, Coves, J. | | Deposit date: | 2014-04-07 | | Release date: | 2014-04-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Crystal Structure of the Anti-Sigma Factor Cnry in Complex with the Sigma Factor Cnrh Shows a New Structural Class of Anti- Sigma Factors Targeting Extracytoplasmic-Function Sigma Factors.
J.Mol.Biol., 426, 2014
|
|
1P80
 
 | | Crystal structure of the D181Q variant of catalase HPII from E. coli | | Descriptor: | Catalase HPII, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Chelikani, P, Carpena, X, Fita, I, Loewen, P.C. | | Deposit date: | 2003-05-06 | | Release date: | 2003-09-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | An electrical potential in the access channel of catalases enhances catalysis
J.Biol.Chem., 278, 2003
|
|
4NDD
 
 | | X-ray structure of a double mutant of calexcitin - a neuronal calcium-signalling protein | | Descriptor: | CALCIUM ION, Calexcitin | | Authors: | Erskine, P.T, Fokas, A, Muriithi, C, Razzall, E, Bowyer, A, Findlow, I.S, Werner, J.M, Wallace, B.A, Wood, S.P, Cooper, J.B. | | Deposit date: | 2013-10-25 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-ray, spectroscopic and normal-mode dynamics of calexcitin: structure-function studies of a neuronal calcium-signalling protein.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8SFJ
 
 | | WT CRISPR-Cas12a with a 10bp R-loop | | Descriptor: | CRISPR-associated endonuclease Cas12a, DNA (5'-D(P*CP*AP*CP*TP*TP*AP*TP*CP*AP*CP*TP*AP*AP*AP*AP*GP*AP*TP*CP*GP*GP*AP*AP*G)-3'), DNA (5'-D(P*CP*TP*TP*CP*CP*GP*AP*TP*CP*TP*TP*TP*TP*AP*GP*TP*GP*AP*T)-3'), ... | | Authors: | Strohkendl, I, Taylor, D.W. | | Deposit date: | 2023-04-11 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | WT CRISPR-Cas12a with a 10bp R-loop
To Be Published
|
|
8SFQ
 
 | |
2IW4
 
 | | CRYSTAL STRUCTURE OF BASILLUS SUBTILIS FAMILY II INORGANIC PYROPHOSPHATASE MUTANT, H98Q, IN COMPLEX WITH PNP | | Descriptor: | FE (III) ION, GLYCEROL, IMIDODIPHOSPHORIC ACID, ... | | Authors: | Fabrichniy, I.P, Lehtio, L, Oksanen, E, Goldman, A. | | Deposit date: | 2006-06-26 | | Release date: | 2006-11-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A Trimetal Site and Substrate Distortion in a Family II Inorganic Pyrophosphatase.
J.Biol.Chem., 282, 2007
|
|
