4WFN
 
 | | Crystal structure of the large ribosomal subunit (50S) of Deinococcus radiodurans containing a three residue insertion in L22 in complex with erythromycin | | 分子名称: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | 著者 | Wekselman, I, Zimmerman, E, Rozenberg, H, Bashan, A, Yonath, A. | | 登録日 | 2014-09-16 | | 公開日 | 2015-12-23 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.54 Å) | | 主引用文献 | The Ribosomal Protein uL22 Modulates the Shape of the Protein Exit Tunnel.
Structure, 25, 2017
|
|
4WEQ
 
 | | Crystal structure of NADPH-dependent glyoxylate/hydroxypyruvate reductase SMc02828 (SmGhrA) from Sinorhizobium meliloti in complex with NADP and sulfate | | 分子名称: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | 著者 | Sroka, P, Gasiorowska, O.A, Handing, K.B, Shabalin, I.G, Osinski, T, Hillerich, B.S, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2014-09-10 | | 公開日 | 2014-09-24 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural, Biochemical, and Evolutionary Characterizations of Glyoxylate/Hydroxypyruvate Reductases Show Their Division into Two Distinct Subfamilies.
Biochemistry, 57, 2018
|
|
4WQD
 
 | | Thiosulfate dehydrogenase (TsdA) from Allochromatium vinosum - K208G mutant | | 分子名称: | 1,2-ETHANEDIOL, GUANIDINE, HEME C, ... | | 著者 | Brito, J.A, Denkmann, K, Pereira, I.A.C, Dahl, C, Archer, M. | | 登録日 | 2014-10-21 | | 公開日 | 2015-02-18 | | 最終更新日 | 2015-04-15 | | 実験手法 | X-RAY DIFFRACTION (1.22 Å) | | 主引用文献 | Thiosulfate Dehydrogenase (TsdA) from Allochromatium vinosum: STRUCTURAL AND FUNCTIONAL INSIGHTS INTO THIOSULFATE OXIDATION.
J.Biol.Chem., 290, 2015
|
|
8UYS
 
 | | SARS-CoV-2 5' proximal stem-loop 5 | | 分子名称: | SARS-CoV-2 RNA SL5 domain. | | 著者 | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | 登録日 | 2023-11-14 | | 公開日 | 2023-12-06 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (4.7 Å) | | 主引用文献 | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UYJ
 
 | | BtCoV-HKU5 5' proximal stem-loop 5, conformation 4 | | 分子名称: | BtCoV-HKU5 5' proximal stem-loop 5, conformation 4 | | 著者 | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | 登録日 | 2023-11-13 | | 公開日 | 2023-12-06 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (7.3 Å) | | 主引用文献 | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UYL
 
 | | MERS 5' proximal stem-loop 5, conformation 2 | | 分子名称: | MERS 5' proximal stem-loop 5 | | 著者 | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | 登録日 | 2023-11-13 | | 公開日 | 2023-12-06 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (6.4 Å) | | 主引用文献 | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UYK
 
 | | MERS 5' proximal stem-loop 5, conformation 1 | | 分子名称: | MERS 5' proximal stem-loop 5 | | 著者 | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | 登録日 | 2023-11-13 | | 公開日 | 2023-12-06 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (6.9 Å) | | 主引用文献 | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UYM
 
 | | MERS 5' proximal stem-loop 5, conformation 3 | | 分子名称: | MERS 5' proximal stem-loop 5 | | 著者 | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | 登録日 | 2023-11-13 | | 公開日 | 2023-12-06 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (6.4 Å) | | 主引用文献 | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UYG
 
 | | BtCoV-HKU5 5' proximal stem-loop 5, conformation 2 | | 分子名称: | RNA (135-MER) | | 著者 | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | 登録日 | 2023-11-13 | | 公開日 | 2023-12-06 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (6.4 Å) | | 主引用文献 | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UYE
 
 | | BtCoV-HKU5 5' proximal stem-loop 5, conformation 1 | | 分子名称: | BtCoV-HKU5 5' proximal stem-loop 5 | | 著者 | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | 登録日 | 2023-11-13 | | 公開日 | 2023-12-06 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (5.9 Å) | | 主引用文献 | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UYP
 
 | | SARS-CoV-1 5' proximal stem-loop 5 | | 分子名称: | SARS-CoV-1 5' proximal stem-loop 5 | | 著者 | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | 登録日 | 2023-11-13 | | 公開日 | 2023-12-20 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (7.1 Å) | | 主引用文献 | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
4WFB
 
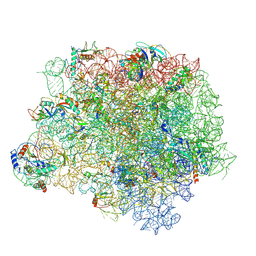 | | The crystal structure of the large ribosomal subunit of Staphylococcus aureus in complex with BC-3205 | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 23S rRNA, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | 著者 | Eyal, Z, Matzov, D, Krupkin, M, Wekselman, I, Zimmerman, E, Rozenberg, H, Bashan, A, Yonath, A.E. | | 登録日 | 2014-09-14 | | 公開日 | 2015-10-21 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.43 Å) | | 主引用文献 | Structural insights into species-specific features of the ribosome from the pathogen Staphylococcus aureus.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4WQA
 
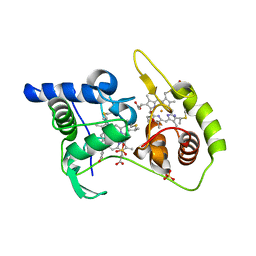 | | Thiosulfate dehydrogenase (TsdA) from Allochromatium vinosum - tetrathionate co-crystallization | | 分子名称: | 1,2-ETHANEDIOL, HEME C, IODIDE ION, ... | | 著者 | Brito, J.A, Denkmann, K, Pereira, I.A.C, Dahl, C, Archer, M. | | 登録日 | 2014-10-21 | | 公開日 | 2015-02-18 | | 最終更新日 | 2015-04-15 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | Thiosulfate Dehydrogenase (TsdA) from Allochromatium vinosum: STRUCTURAL AND FUNCTIONAL INSIGHTS INTO THIOSULFATE OXIDATION.
J.Biol.Chem., 290, 2015
|
|
4WQ8
 
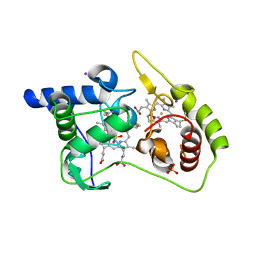 | | Thiosulfate dehydrogenase (TsdA) from Allochromatium vinosum - tetrathionate soak | | 分子名称: | HEME C, IODIDE ION, SULFATE ION, ... | | 著者 | Brito, J.A, Denkmann, K, Pereira, I.A.C, Dahl, C, Archer, M. | | 登録日 | 2014-10-21 | | 公開日 | 2015-02-18 | | 最終更新日 | 2018-03-07 | | 実験手法 | X-RAY DIFFRACTION (1.3989 Å) | | 主引用文献 | Thiosulfate Dehydrogenase (TsdA) from Allochromatium vinosum: STRUCTURAL AND FUNCTIONAL INSIGHTS INTO THIOSULFATE OXIDATION.
J.Biol.Chem., 290, 2015
|
|
4WPH
 
 | |
7BGI
 
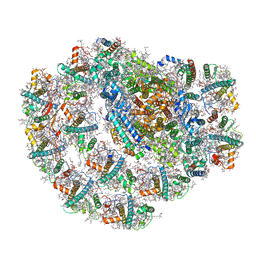 | | Photosystem I of a temperature sensitive mutant Chlamydomonas reinhardtii | | 分子名称: | (1~{S})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(4~{S})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15,17-nonaenyl]cyclohex-3-en-1-ol, (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, (2S)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-hydroxypropyl hexadecanoate, ... | | 著者 | Caspy, I, Nelson, N. | | 登録日 | 2021-01-07 | | 公開日 | 2021-12-08 | | 最終更新日 | 2021-12-15 | | 実験手法 | ELECTRON MICROSCOPY (2.54 Å) | | 主引用文献 | Dimeric and high-resolution structures of Chlamydomonas Photosystem I from a temperature-sensitive Photosystem II mutant
Commun Biol, 4, 2021
|
|
7BLX
 
 | | Photosystem I of a temperature sensitive mutant Chlamydomonas reinhardtii | | 分子名称: | (1~{S})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(4~{S})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15,17-nonaenyl]cyclohex-3-en-1-ol, (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, (2S)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-hydroxypropyl hexadecanoate, ... | | 著者 | Caspy, I, Nelson, N. | | 登録日 | 2021-01-19 | | 公開日 | 2021-12-15 | | 実験手法 | ELECTRON MICROSCOPY (3.15 Å) | | 主引用文献 | Dimeric and high-resolution structures of Chlamydomonas Photosystem I from a temperature-sensitive Photosystem II mutant
Commun Biol, 4, 2021
|
|
9B63
 
 | |
9FST
 
 | | Yeast 20S proteasome with human beta1i (1-51) in complex with epoxyketone inhibitor LU-001i | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | 著者 | Maurits, E, Huber, E.M, Dekker, P.M, Wang, X, Heinemeyer, W, Florea, B.I, Groll, M, Overkleeft, H.S. | | 登録日 | 2024-06-21 | | 公開日 | 2024-07-17 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structure-based design of peptide epoxyketones selectively targeting the three human immunoproteasome active sites
To Be Published
|
|
9B5Z
 
 | |
9B64
 
 | |
9GG1
 
 | | P301T type I tau filaments from human brain | | 分子名称: | Isoform Tau-D of Microtubule-associated protein tau | | 著者 | Schweighauser, M, Shi, Y, Murzin, A.G, Garringer, H.J, Vidal, R, Murrell, J.R, Erro, M.E, Seelaar, H, Ferrer, I, van Swieten, J.C, Ghetti, B, Scheres, S.H.W, Goedert, M. | | 登録日 | 2024-08-12 | | 公開日 | 2024-09-11 | | 実験手法 | ELECTRON MICROSCOPY (2.26 Å) | | 主引用文献 | Novel tau filament folds in individuals with MAPT mutations P301L and P301T.
Biorxiv, 2024
|
|
9B60
 
 | |
9B61
 
 | |
9B6A
 
 | |
