2IKO
 
 | |
9B64
 
 | |
1Y9L
 
 | | The X-ray structure of an secretion system protein | | 分子名称: | ACETATE ION, Lipoprotein mxiM, UNDECANE | | 著者 | Lario, P.I, Pfuetzer, R.A, Frey, E.A, Creagh, L, Haynes, C, Maurelli, A.T, Strynadka, N.C. | | 登録日 | 2004-12-15 | | 公開日 | 2005-04-12 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structure and biochemical analysis of a secretin pilot protein.
Embo J., 24, 2005
|
|
2IT6
 
 | | Crystal Structure of DCSIGN-CRD with man2 | | 分子名称: | CALCIUM ION, CD209 antigen, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose | | 著者 | Weis, W.I, Feinberg, H, Castelli, R, Drickamer, K, Seeberger, P.H. | | 登録日 | 2006-10-19 | | 公開日 | 2006-12-05 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Multiple modes of binding enhance the affinity of DC-SIGN for high mannose N-linked glycans found on viral glycoproteins.
J.Biol.Chem., 282, 2007
|
|
1Y9T
 
 | |
9PAP
 
 | |
9B63
 
 | |
9B5Z
 
 | |
9FT1
 
 | | Yeast 20S proteasome in complex with epoxyketone inhibitor 13 | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | 著者 | Maurits, E, Huber, E.M, Dekker, P.M, Wang, X, Heinemeyer, W, Florea, B.I, Groll, M, Overkleeft, H.S. | | 登録日 | 2024-06-23 | | 公開日 | 2024-07-10 | | 最終更新日 | 2024-07-17 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure-based design of peptide epoxyketones selectively targeting the three human immunoproteasome active sites
to be published
|
|
9B00
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with berberine analog of chloramphenicol CAM-BER, mRNA, deacylated A- and E-site tRNAphe, and deacylated P-site tRNAmet at 2.80A resolution | | 分子名称: | 13-(2-{[(1R,2R)-1,3-dihydroxy-1-(4-nitrophenyl)propan-2-yl]amino}-2-oxoethyl)-9,10-dimethoxy-5,6-dihydro-2H-[1,3]dioxolo[4,5-g]isoquinolino[3,2-a]isoquinolin-7-ium, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | 著者 | Batool, Z, Pavlova, J.A, Paranjpe, M.N, Tereshchenkov, A.G, Lukianov, D.A, Osterman, I.A, Bogdanov, A.A, Sumbatyan, N.V, Polikanov, Y.S. | | 登録日 | 2024-03-11 | | 公開日 | 2024-08-07 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Berberine analog of chloramphenicol exhibits a distinct mode of action and unveils ribosome plasticity.
Structure, 2024
|
|
9MSI
 
 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 T18N | | 分子名称: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | 著者 | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | 登録日 | 1999-01-24 | | 公開日 | 1999-04-28 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
8WB8
 
 | |
2J5D
 
 | | NMR structure of BNIP3 transmembrane domain in lipid bicelles | | 分子名称: | BCL2/ADENOVIRUS E1B 19 KDA PROTEIN-INTERACTING PROTEIN 3 | | 著者 | Bocharov, E.V, Pustovalova, Y.E, Volynsky, P.E, Maslennikov, I.V, Goncharuk, M.V, Ermolyuk, Y.S, Arseniev, A.S. | | 登録日 | 2006-09-14 | | 公開日 | 2007-04-17 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Unique dimeric structure of BNip3 transmembrane domain suggests membrane permeabilization as a cell death trigger.
J. Biol. Chem., 282, 2007
|
|
9B60
 
 | |
1JVR
 
 | | STRUCTURE OF THE HTLV-II MATRIX PROTEIN, NMR, 20 STRUCTURES | | 分子名称: | HUMAN T-CELL LEUKEMIA VIRUS TYPE II MATRIX PROTEIN | | 著者 | Christensen, A.M, Massiah, M.A, Turner, B.G, Sundquist, W.I, Summers, M.F. | | 登録日 | 1996-10-03 | | 公開日 | 1997-03-12 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Three-dimensional structure of the HTLV-II matrix protein and comparative analysis of matrix proteins from the different classes of pathogenic human retroviruses.
J.Mol.Biol., 264, 1996
|
|
9B61
 
 | |
1JSA
 
 | | MYRISTOYLATED RECOVERIN WITH TWO CALCIUMS BOUND, NMR, 24 STRUCTURES | | 分子名称: | CALCIUM ION, MYRISTIC ACID, RECOVERIN | | 著者 | Ames, J.B, Ishima, R, Tanaka, T, Gordon, J.I, Stryer, L, Ikura, M. | | 登録日 | 1997-06-04 | | 公開日 | 1997-10-15 | | 最終更新日 | 2022-02-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Molecular mechanics of calcium-myristoyl switches.
Nature, 389, 1997
|
|
9FT0
 
 | | Yeast 20S proteasome in complex with epoxyketone inhibitor 42 | | 分子名称: | (2S)-N-[(2S)-1-[[(1S)-2-cyclohexyl-1-[(2R,3S,6R,7S)-3-methanoyl-2,6-dimethyl-6,7-bis(oxidanyl)-1,4-oxazepan-7-yl]ethyl]amino]-3-(4-methoxyphenyl)-1-oxidanylidene-propan-2-yl]-2-(2-morpholin-4-ylethanoylamino)-4-oxidanyl-butanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | 著者 | Maurits, E, Huber, E.M, Dekker, P.M, Wang, X, Heinemeyer, W, Florea, B.I, Groll, M, Overkleeft, H.S. | | 登録日 | 2024-06-23 | | 公開日 | 2024-07-10 | | 最終更新日 | 2024-07-17 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structure-based design of peptide epoxyketones selectively targeting the three human immunoproteasome active sites
to be published
|
|
9B67
 
 | |
1K5H
 
 | | 1-deoxy-D-xylulose-5-phosphate reductoisomerase | | 分子名称: | 1-deoxy-D-xylulose-5-phosphate reductoisomerase | | 著者 | Reuter, K, Sanderbrand, S, Jomaa, H, Wiesner, J, Steinbrecher, I, Beck, E, Hintz, M, Klebe, G, Stubbs, M.T. | | 登録日 | 2001-10-10 | | 公開日 | 2002-02-27 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of 1-deoxy-D-xylulose-5-phosphate reductoisomerase, a crucial enzyme in the non-mevalonate pathway of isoprenoid biosynthesis.
J.Biol.Chem., 277, 2002
|
|
1K7E
 
 | | CRYSTAL STRUCTURE OF WILD-TYPE TRYPTOPHAN SYNTHASE COMPLEXED WITH N-[1H-INDOL-3-YL-ACETYL]GLYCINE ACID | | 分子名称: | N-[1H-INDOL-3-YL-ACETYL]GLYCINE ACID, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, ... | | 著者 | Weyand, M, Schlichting, I, Marabotti, A, Mozzarelli, A. | | 登録日 | 2001-10-19 | | 公開日 | 2002-07-10 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structures of a new class of allosteric effectors complexed to tryptophan synthase.
J.Biol.Chem., 277, 2002
|
|
9FSV
 
 | | Yeast 20S proteasome with human beta2i (1-53) in complex with epoxyketone inhibitor 42 | | 分子名称: | (2S)-N-[(2S)-1-[[(1S)-2-cyclohexyl-1-[(2R,3S,6R,7S)-3-methanoyl-2,6-dimethyl-6,7-bis(oxidanyl)-1,4-oxazepan-7-yl]ethyl]amino]-3-(4-methoxyphenyl)-1-oxidanylidene-propan-2-yl]-2-(2-morpholin-4-ylethanoylamino)-4-oxidanyl-butanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | 著者 | Maurits, E, Huber, E.M, Dekker, P.M, Wang, X, Heinemeyer, W, Florea, B.I, Groll, M, Overkleeft, H.S. | | 登録日 | 2024-06-22 | | 公開日 | 2024-07-17 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structure-based design of peptide epoxyketones selectively targeting the three human immunoproteasome active sites
to be published
|
|
1K8Z
 
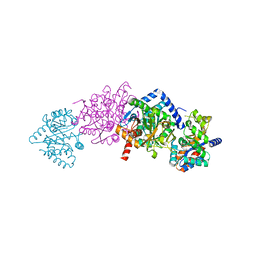 | | CRYSTAL STRUCTURE OF THE TRYPTOPHAN SYNTHASE BETA-SER178PRO MUTANT COMPLEXED WITH N-[1H-INDOL-3-YL-ACETYL]GLYCINE ACID | | 分子名称: | N-[1H-INDOL-3-YL-ACETYL]GLYCINE ACID, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, ... | | 著者 | Weyand, M, Schlichting, I, Marabotti, A, Mozzarelli, A. | | 登録日 | 2001-10-26 | | 公開日 | 2002-06-19 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of the beta Ser178--> Pro mutant of tryptophan synthase. A "knock-out" allosteric enzyme.
J.Biol.Chem., 277, 2002
|
|
2J8G
 
 | |
1K3O
 
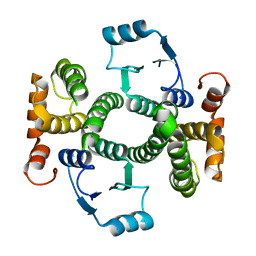 | | Crystal Structure Analysis of apo Glutathione S-Transferase | | 分子名称: | GLUTATHIONE S-TRANSFERASE A1 | | 著者 | Le Trong, I, Stenkamp, R.E, Ibarra, C, Atkins, W.M, Adman, E.T. | | 登録日 | 2001-10-03 | | 公開日 | 2002-10-30 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | 1.3-A resolution structure of human glutathione S-transferase with S-hexyl glutathione bound reveals possible extended ligandin binding site.
Proteins, 48, 2002
|
|
