2QGC
 
 | |
5MWJ
 
 | | Structure Enabled Discovery of a Stapled Peptide Inhibitor to Target the Oncogenic Transcriptional Repressor TLE1 | | Descriptor: | DIMETHYL SULFOXIDE, Transducin-like enhancer protein 1, pepide inhibtor | | Authors: | McGrath, S, Tortorici, M, Vidler, L, Drouin, L, Westwood, I, Gimeson, P, Van Montfort, R, Hoelder, S. | | Deposit date: | 2017-01-18 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure-Enabled Discovery of a Stapled Peptide Inhibitor to Target the Oncogenic Transcriptional Repressor TLE1.
Chemistry, 23, 2017
|
|
2LCP
 
 | | NMR structure of calcium loaded, un-myristoylated human NCS-1 | | Descriptor: | CALCIUM ION, Neuronal calcium sensor 1 | | Authors: | Heidarsson, P.O, Bjerrum-Bohr, I.J, Bellucci, L, Gitte, J, Corni, S, Poulsen, F.M, Finn, B.E, Di Felice, R, Kragelund, B. | | Deposit date: | 2011-05-05 | | Release date: | 2012-02-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The C-terminal tail of human neuronal calcium sensor 1 regulates the conformational stability of the ca(2+)-activated state.
J.Mol.Biol., 417, 2012
|
|
2QI2
 
 | | Crystal structure of the Thermoplasma acidophilum Pelota protein | | Descriptor: | Cell division protein pelota related protein | | Authors: | Lee, H.H, Kim, Y.S, Kim, K.H, Heo, I.H, Kim, S.K, Kim, O, Suh, S.W. | | Deposit date: | 2007-07-03 | | Release date: | 2007-10-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and functional insights into dom34, a key component of no-go mRNA decay
Mol.Cell, 27, 2007
|
|
5MYF
 
 | | Convergent evolution involving dimeric and trimeric dUTPases in signalling. | | Descriptor: | dUTPase from DI S. aureus phage | | Authors: | Donderis, J, Bowring, J, Maiques, E, Ciges-Tomas, J.R, Alite, C, Mehmedov, I, Tormo-Mas, M.A, Penades, J.R, Marina, A. | | Deposit date: | 2017-01-26 | | Release date: | 2017-09-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Convergent evolution involving dimeric and trimeric dUTPases in pathogenicity island mobilization.
PLoS Pathog., 13, 2017
|
|
2LKM
 
 | |
5MTZ
 
 | | Crystal structure of a long form RNase Z from yeast | | Descriptor: | PHOSPHATE ION, Ribonuclease Z, ZINC ION | | Authors: | Li de la Sierra-Gallay, I, Miao, M, van Tilbeurgh, H. | | Deposit date: | 2017-01-11 | | Release date: | 2017-06-21 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | The crystal structure of Trz1, the long form RNase Z from yeast.
Nucleic Acids Res., 45, 2017
|
|
5MUI
 
 | | Glycoside hydrolase BT_0996 | | Descriptor: | Beta-galactosidase, beta-L-arabinofuranose-(1-2)-alpha-L-rhamnopyranose-(1-2)-[alpha-L-rhamnopyranose-(1-3)]alpha-L-arabinopyranose-(1-4)-[4-O-[(1R)-1-hydroxyethyl]-2-O-methyl-alpha-L-fucopyranose-(1-2)]beta-D-galactopyranose-(1-2)-alpha-D-aceric acid-(1-3)-alpha-L-rhamnopyranose | | Authors: | Basle, A, Ndeh, D, Rogowski, A, Cartmell, A, Luis, A.S, Venditto, I, Labourel, A, Gilbert, H.J. | | Deposit date: | 2017-01-13 | | Release date: | 2017-03-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Complex pectin metabolism by gut bacteria reveals novel catalytic functions.
Nature, 544, 2017
|
|
2LLX
 
 | |
3KTL
 
 | | Crystal Structure of an I71A human GSTA1-1 mutant in complex with S-hexylglutathione | | Descriptor: | Glutathione S-transferase A1, S-HEXYLGLUTATHIONE | | Authors: | Achilonu, I.A, Gildenhuys, S, Fernandes, M.A, Fanucchi, S, Dirr, H.W. | | Deposit date: | 2009-11-25 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The role of a topologically conserved isoleucine in glutathione transferase structure, stability and function.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2L10
 
 | | Solution Structure of the R6 Domain of Talin | | Descriptor: | Talin-1 | | Authors: | Goult, B.T, Gingras, A.R, Bate, N, Roberts, G.C.K, Barsukov, I.L, Critchley, D.R. | | Deposit date: | 2010-07-22 | | Release date: | 2011-08-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | RIAM and vinculin binding to talin are mutually exclusive and regulate adhesion assembly and turnover.
J.Biol.Chem., 288, 2013
|
|
1DSJ
 
 | | NMR SOLUTION STRUCTURE OF VPR50_75, 20 STRUCTURES | | Descriptor: | VPR PROTEIN | | Authors: | Yao, S, Torres, A.M, Azad, A.A, Macreadie, I.G, Norton, R.S. | | Deposit date: | 1997-10-23 | | Release date: | 1998-07-01 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Helical structure of polypeptides from the C-terminal half of HIV-1 VPR.
Protein Pept.Lett., 5, 1998
|
|
1DBB
 
 | |
5MSI
 
 | |
2L38
 
 | | R29Q Sticholysin II mutant | | Descriptor: | Sticholysin-2 | | Authors: | Castrillo, I, Alegre-Cebollada, J, Martinez-del-Pozo, A, Gavilanes, J, Bruix, M. | | Deposit date: | 2010-09-10 | | Release date: | 2010-09-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of StnIIR29Q, a defective lipid binding mutant of the sea anemone actinoporin Sticholysin II
To be Published
|
|
1D9S
 
 | | TUMOR SUPPRESSOR P15(INK4B) STRUCTURE BY COMPARATIVE MODELING AND NMR DATA | | Descriptor: | CYCLIN-DEPENDENT KINASE 4 INHIBITOR B | | Authors: | Yuan, C, Ji, L, Selby, T.L, Byeon, I.J.L, Tsai, M.D. | | Deposit date: | 1999-10-29 | | Release date: | 2000-07-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Tumor suppressor INK4: comparisons of conformational properties between p16(INK4A) and p18(INK4C).
J.Mol.Biol., 294, 1999
|
|
5MNW
 
 | |
5MJX
 
 | | 2'F-ANA/DNA Chimeric TBA Quadruplex structure | | Descriptor: | DNA (5'-D(*GP*GP*(FT)P*TP*GP*GP*TP*GP*TP*GP*GP*TP*TP*GP*G)-3') | | Authors: | Lietard, J, Abou Assi, H, Gomez-Pinto, I, Gonzalez, C, Somoza, M.M, Damha, M.J. | | Deposit date: | 2016-12-02 | | Release date: | 2017-02-01 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Mapping the affinity landscape of Thrombin-binding aptamers on 2 F-ANA/DNA chimeric G-Quadruplex microarrays.
Nucleic Acids Res., 45, 2017
|
|
5MWW
 
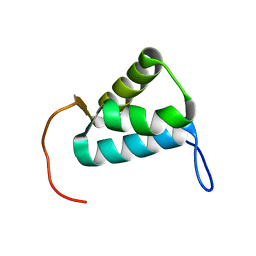 | | Sigma1.1 domain of sigmaA from Bacillus subtilis | | Descriptor: | RNA polymerase sigma factor SigA | | Authors: | Zachrdla, M, Padrta, P, Rabatinova, A, Sanderova, H, Barvik, I, Krasny, L, Zidek, L. | | Deposit date: | 2017-01-20 | | Release date: | 2017-06-14 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Solution structure of domain 1.1 of the sigma (A) factor from Bacillus subtilis is preformed for binding to the RNA polymerase core.
J. Biol. Chem., 292, 2017
|
|
4Y1B
 
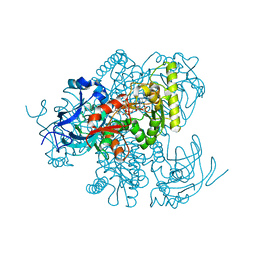 | |
5N49
 
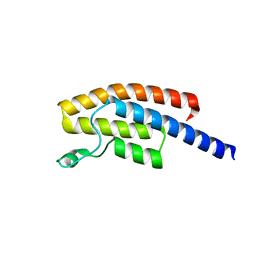 | | BRPF2 in complex with Compound 7 | | Descriptor: | 2-(1,3,6-trimethyl-2-oxidanylidene-benzimidazol-5-yl)benzo[de]isoquinoline-1,3-dione, Bromodomain-containing protein 1 | | Authors: | Bouche, L, Christ, C.D, Siegel, S, Fernandez-Montalvan, A.E, Holton, S.J, Fedorov, O, ter Laak, A, Sugawara, T, Stoeckigt, D, Tallant, C, Bennett, J, Monteiro, O, Saez, L.D, Siejka, P, Meier, J, Puetter, V, Weiske, J, Mueller, S, Huber, K.V.M, Hartung, I.V, Haendler, B. | | Deposit date: | 2017-02-10 | | Release date: | 2017-05-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Benzoisoquinolinediones as Potent and Selective Inhibitors of BRPF2 and TAF1/TAF1L Bromodomains.
J. Med. Chem., 60, 2017
|
|
2L4O
 
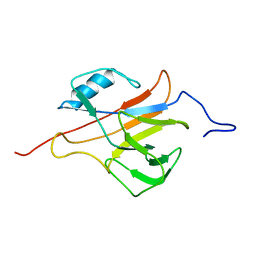 | | Solution structure of the Streptococcus pneumoniae RrgB pilus backbone D1 domain | | Descriptor: | Cell wall surface anchor family protein | | Authors: | Gentile, M.A, Melchiorre, S, Emolo, C, Moschioni, M, Gianfaldoni, C, Pancotto, L, Ferlenghi, I, Scarselli, M, Pansegrau, W, Veggi, D, Merola, M, Cantini, F, Ruggiero, P, Banci, L, Masignani, V. | | Deposit date: | 2010-10-11 | | Release date: | 2011-03-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Streptococcus pneumoniae RrgB pilus backbone D1 domain: in vivo protection and implications for the molecular mechanisms of pilus polymerization
To be Published
|
|
2LOM
 
 | | Backbone structure of human membrane protein HIGD1A | | Descriptor: | HIG1 domain family member 1A | | Authors: | Blain, K, Klammt, C, Maslennikov, I, Kwiatkowski, W, Choe, S. | | Deposit date: | 2012-01-26 | | Release date: | 2012-05-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Facile backbone structure determination of human membrane proteins by NMR spectroscopy.
Nat.Methods, 9, 2012
|
|
1DAB
 
 | |
2LOS
 
 | | Backbone structure of human membrane protein TMEM14C | | Descriptor: | Transmembrane protein 14C | | Authors: | Klammt, C, Vajpai, N, Maslennikov, I, Riek, R, Choe, S. | | Deposit date: | 2012-01-26 | | Release date: | 2012-05-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Facile backbone structure determination of human membrane proteins by NMR spectroscopy.
Nat.Methods, 9, 2012
|
|
