5BZW
 
 | |
1LZI
 
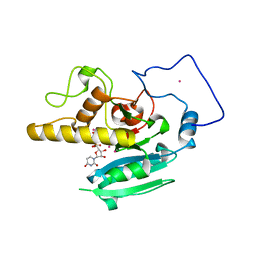 | | Glycosyltransferase A + UDP + H antigen acceptor | | Descriptor: | Glycosyltransferase A, MANGANESE (II) ION, MERCURY (II) ION, ... | | Authors: | Patenaude, S.I, Seto, N.O.L, Borisova, S.N, Szpacenko, A, Marcus, S.L, Palcic, M.M, Evans, S.V. | | Deposit date: | 2002-06-10 | | Release date: | 2002-08-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The structural basis for specificity
in human ABO(H) blood group biosynthesis.
Nat.Struct.Biol., 9, 2002
|
|
5TC0
 
 | |
6YZ7
 
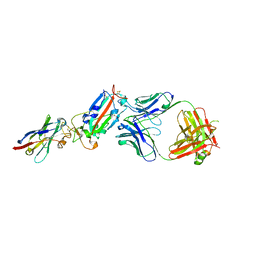 | | H11-D4, SARS-CoV-2 RBD, CR3022 ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody Cr3022, Antibody light chain, ... | | Authors: | Naismith, J.H, Ren, J, Zhou, D, Zhao, Y, Stuart, D.I. | | Deposit date: | 2020-05-06 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural characterisation of a nanobody derived from a naive library that neutralises SARS-CoV-2
To Be Published, 2020
|
|
5NOY
 
 | | Structure of cyclophilin A in complex with 3,4-diaminobenzamide | | Descriptor: | 3,4-bis(azanyl)benzamide, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Georgiou, C, Mcnae, I.W, Ioannidis, H, Julien, M, Walkinshaw, M.D. | | Deposit date: | 2017-04-13 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Pushing the Limits of Detection of Weak Binding Using Fragment-Based Drug Discovery: Identification of New Cyclophilin Binders.
J. Mol. Biol., 429, 2017
|
|
8ATJ
 
 | | Crystal Structure of Shank2-SAM domain | | Descriptor: | CHLORIDE ION, FORMIC ACID, Isoform 4 of SH3 and multiple ankyrin repeat domains protein 2, ... | | Authors: | Bento, I, Gracia Alai, M, Kreienkamp, J.-H. | | Deposit date: | 2022-08-23 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.117 Å) | | Cite: | Structural deficits in key domains of Shank2 lead to alterations in postsynaptic nanoclusters and to a neurodevelopmental disorder in humans.
Mol Psychiatry, 2022
|
|
5TD2
 
 | |
6RZY
 
 | |
8AKO
 
 | | Structure of EspB-EspK complex: the non-identical twin of the PE-PPE-EspG secretion mechanism. | | Descriptor: | ESX-1 secretion-associated protein EspB, ESX-1 secretion-associated protein EspK | | Authors: | Gijsbers, A, Eymery, M, Menart, I, Vinciauskaite, V, Gao, Y, Siliqi, D, Peters, P, Mccarthy, A, Ravelli, R.B.G. | | Deposit date: | 2022-07-30 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.293 Å) | | Cite: | The crystal structure of the EspB-EspK virulence factor-chaperone complex suggests an additional type VII secretion mechanism in Mycobacterium tuberculosis.
J.Biol.Chem., 299, 2022
|
|
2NNR
 
 | | Crystal structure of chagasin, cysteine protease inhibitor from Trypanosoma cruzi | | Descriptor: | CHLORIDE ION, Chagasin, GLYCEROL, ... | | Authors: | Redzynia, I, Bujacz, G, Ljunggren, A, Jaskolski, M, Abrahamson, M. | | Deposit date: | 2006-10-24 | | Release date: | 2007-07-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the parasite protease inhibitor chagasin in complex with a host target cysteine protease
J.Mol.Biol., 371, 2007
|
|
6E85
 
 | | 1.25 Angstrom Resolution Crystal Structure of 4-hydroxythreonine-4-phosphate Dehydrogenase from Klebsiella pneumoniae. | | Descriptor: | CHLORIDE ION, D-threonate 4-phosphate dehydrogenase, FORMIC ACID, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Endres, M, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-07-27 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
6ZHI
 
 | |
5F91
 
 | | Fumarate hydratase of Mycobacterium tuberculosis in complex with formate and allosteric modulator (N-(5-(azepan-1-ylsulfonyl)-2-methoxyphenyl)-2-(4-oxo-3,4-dihydrophthalazin-1-yl)acetamide) | | Descriptor: | CHLORIDE ION, FORMIC ACID, Fumarate hydratase class II, ... | | Authors: | Kasbekar, M, Fischer, G, Mott, B.T, Yasgar, A, Hyvonen, M, Boshoff, H.I, Abell, C, Barry, C.E, Thomas, C.J. | | Deposit date: | 2015-12-09 | | Release date: | 2016-06-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Selective small molecule inhibitor of the Mycobacterium tuberculosis fumarate hydratase reveals an allosteric regulatory site.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5C9H
 
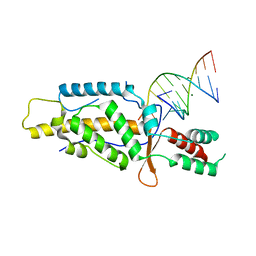 | | Structural Basis of Template Boundary Definition in Tetrahymena Telomerase | | Descriptor: | MAGNESIUM ION, RNA (5'-R(P*AP*GP*AP*AP*CP*UP*GP*UP*CP*A)-3'), RNA (5'-R(P*UP*CP*AP*UP*UP*CP*AP*GP*UP*UP*CP*U)-3'), ... | | Authors: | Jansson, L.I, Akiyama, B.M, Ooms, A, Lu, C, Rubin, S.M, Stone, M.D. | | Deposit date: | 2015-06-26 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of template-boundary definition in Tetrahymena telomerase.
Nat.Struct.Mol.Biol., 22, 2015
|
|
1P2Q
 
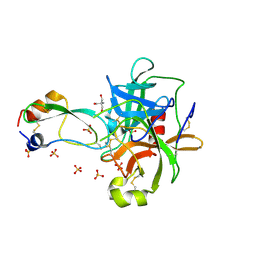 | | Structural consequences of accommodation of four non-cognate amino-acid residues in the S1 pocket of bovine trypsin and chymotrypsin | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Chymotrypsinogen A, Pancreatic trypsin inhibitor, ... | | Authors: | Helland, R, Czapinska, H, Leiros, I, Olufsen, M, Otlewski, J, Smalaas, A.O. | | Deposit date: | 2003-04-15 | | Release date: | 2004-04-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural consequences of accommodation of four non-cognate amino acid residues in the S1 pocket of bovine trypsin and chymotrypsin.
J.Mol.Biol., 333, 2003
|
|
6EMF
 
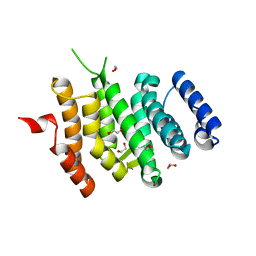 | |
4N34
 
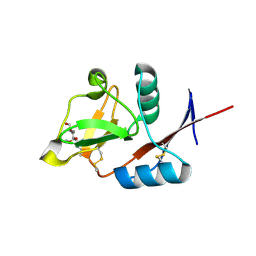 | | Structure of langerin CRD I313 with alpha-MeGlcNAc | | Descriptor: | C-type lectin domain family 4 member K, CALCIUM ION, methyl 2-acetamido-2-deoxy-alpha-D-glucopyranoside | | Authors: | Feinberg, H, Rowntree, T.J.W, Tan, S.L.W, Drickamer, K, Weis, W.I, Taylor, M.E. | | Deposit date: | 2013-10-06 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Common polymorphisms in human langerin change specificity for glycan ligands.
J.Biol.Chem., 288, 2013
|
|
1P4R
 
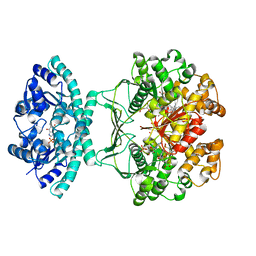 | | Crystal Structure of Human ATIC in complex with folate-based inhibitor BW1540U88UD | | Descriptor: | AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, Bifunctional purine biosynthesis protein PURH, N-[(S)-(4-{[(2-AMINO-4-HYDROXYQUINAZOLIN-6-YL)(DIHYDROXY)-LAMBDA~4~-SULFANYL]AMINO}PHENYL)(HYDROXY)METHYL]-L-GLUTAMIC ACID, ... | | Authors: | Cheong, C.-G, Greasley, S.E, Horton, P.A, Beardsley, G.P, Wilson, I.A. | | Deposit date: | 2003-04-23 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structures of human bifunctional enzyme aminoimidazole-4-carboxamide ribonucleotide transformylase/IMP cyclohydrolase in complex with potent sulfonyl-containing antifolates
J.Biol.Chem., 279, 2004
|
|
5NKO
 
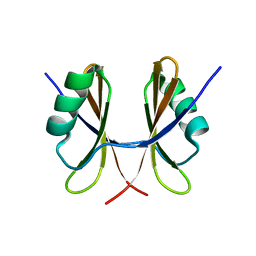 | | Solution structure of the C-terminal domain of S. aureus Hibernating Promoting Factor (CTD-SaHPF) | | Descriptor: | Ribosome hibernation promotion factor | | Authors: | Usachev, K.S, Khusainov, I.S, Ayupov, R.K, Validov, S.Z, Kieffer, B, Yusupov, M.M. | | Deposit date: | 2017-03-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structures and dynamics of hibernating ribosomes from Staphylococcus aureus mediated by intermolecular interactions of HPF.
EMBO J., 36, 2017
|
|
5CEA
 
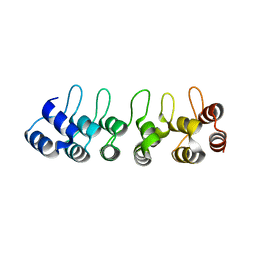 | |
8J3T
 
 | | Complex structure of human cytomegalovirus protease and a non-covalent small-molecule ligand | | Descriptor: | (4R)-1-[1-[(S)-[1-cyclopentyl-3-(2-methylphenyl)pyrazol-4-yl]-(4-methylphenyl)methyl]-2-oxidanylidene-pyridin-3-yl]-3-methyl-2-oxidanylidene-N-(3-oxidanylidene-2-azabicyclo[2.2.2]octan-4-yl)imidazolidine-4-carboxamide, Assemblin | | Authors: | Yoshida, S, Sako, Y, Nikaido, E, Ueda, T, Kozono, I, Ichihashi, Y, Nakahashi, A, Onishi, M, Yamatsu, Y, Kato, T, Nishikawa, J, Tachibana, Y. | | Deposit date: | 2023-04-18 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Peptide-to-Small Molecule: Discovery of Non-Covalent, Active-Site Inhibitors of beta-Herpesvirus Proteases.
Acs Med.Chem.Lett., 14, 2023
|
|
1UDY
 
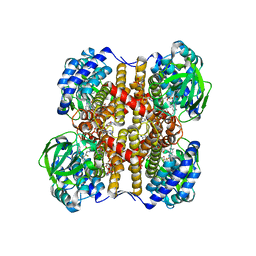 | | Medium-Chain Acyl-CoA Dehydrogenase with 3-Thiaoctanoyl-CoA | | Descriptor: | 3-THIAOCTANOYL-COENZYME A, Acyl-CoA dehydrogenase, medium-chain specific, ... | | Authors: | Satoh, A, Nakajima, Y, Miyahara, I, Hirotsu, K, Tanaka, T, Nishina, Y, Shiga, K, Tamaoki, H, Setoyama, C, Miura, R. | | Deposit date: | 2003-05-07 | | Release date: | 2003-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the transition state analog of medium-chain acyl-CoA dehydrogenase. Crystallographic and molecular orbital studies on the charge-transfer complex of medium-chain acyl-CoA dehydrogenase with 3-thiaoctanoyl-CoA
J.BIOCHEM.(TOKYO), 134, 2003
|
|
8AW4
 
 | |
4C4G
 
 | | Structure-based design of orally bioavailable pyrrolopyridine inhibitors of the mitotic kinase MPS1 | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, DUAL SPECIFICITY PROTEIN KINASE TTK, ... | | Authors: | Naud, S, Westwood, I.M, Faisal, A, Sheldrake, P, Bavetsias, V, Atrash, B, Liu, M, Hayes, A, Schmitt, J, Wood, A, Choi, V, Boxall, K, Mak, G, Gurden, M, Valenti, M, de Haven Brandon, A, Henley, A, Baker, R, McAndrew, C, Matijssen, B, Burke, R, Eccles, S.A, Raynaud, F.I, Linardopoulos, S, van Montfort, R, Blagg, J. | | Deposit date: | 2013-09-05 | | Release date: | 2013-12-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure-Based Design of Orally Bioavailable 1H-Pyrrolo[3, 2-C]Pyridine Inhibitors of the Mitotic Kinase Monopolar Spindle 1 (Mps1).
J.Med.Chem., 56, 2013
|
|
2KOD
 
 | | A high-resolution NMR structure of the dimeric C-terminal domain of HIV-1 CA | | Descriptor: | HIV-1 CA C-terminal domain | | Authors: | Byeon, I.-J.L, Jung, J, Ahn, J, concel, J, Gronenborn, A.M. | | Deposit date: | 2009-09-18 | | Release date: | 2009-11-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural convergence between Cryo-EM and NMR reveals intersubunit interactions critical for HIV-1 capsid function.
Cell(Cambridge,Mass.), 139, 2009
|
|
