3PFQ
 
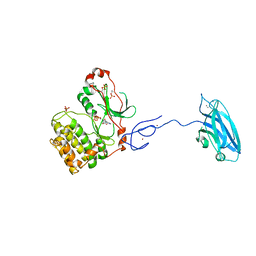 | | Crystal Structure and Allosteric Activation of Protein Kinase C beta II | | Descriptor: | CALCIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Protein kinase C beta type, ... | | Authors: | Leonard, T.A, Rozycki, B, Saidi, L.F, Hummer, G, Hurley, J.H. | | Deposit date: | 2010-10-28 | | Release date: | 2011-02-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Crystal Structure and Allosteric Activation of Protein Kinase C beta II
Cell(Cambridge,Mass.), 144, 2011
|
|
3R42
 
 | |
1AB8
 
 | |
3R3Q
 
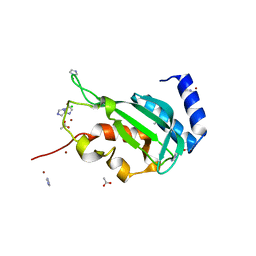 | | Crystal structure of the yeast Vps23 UEV domain | | Descriptor: | ACETATE ION, CHLORIDE ION, IMIDAZOLE, ... | | Authors: | Ren, X, Hurley, J.H. | | Deposit date: | 2011-03-16 | | Release date: | 2011-05-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural basis for endosomal recruitment of ESCRT-I by ESCRT-0 in yeast.
Embo J., 30, 2011
|
|
1BO1
 
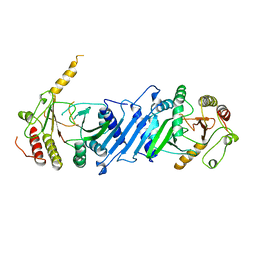 | | PHOSPHATIDYLINOSITOL PHOSPHATE KINASE TYPE II BETA | | Descriptor: | PROTEIN (PHOSPHATIDYLINOSITOL PHOSPHATE KINASE IIBETA) | | Authors: | Rao, V.D, Misra, S, Boronenkov, I.V, Anderson, R.A, Hurley, J.H. | | Deposit date: | 1998-08-02 | | Release date: | 1998-10-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of type IIbeta phosphatidylinositol phosphate kinase: a protein kinase fold flattened for interfacial phosphorylation.
Cell(Cambridge,Mass.), 94, 1998
|
|
3OBQ
 
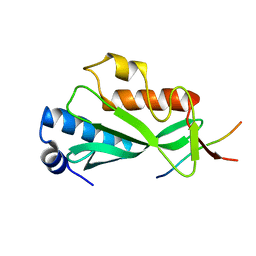 | |
3OBX
 
 | |
3OBS
 
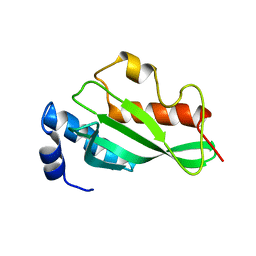 | | Crystal structure of Tsg101 UEV domain | | Descriptor: | Tumor susceptibility gene 101 protein | | Authors: | Im, Y.J, Hurley, J.H. | | Deposit date: | 2010-08-09 | | Release date: | 2010-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystallographic and Functional Analysis of the ESCRT-I /HIV-1 Gag PTAP Interaction.
Structure, 18, 2010
|
|
1ZHX
 
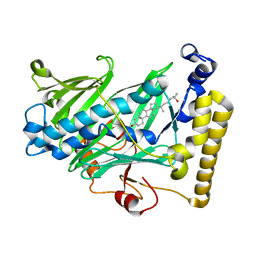 | | Structure of yeast oxysterol binding protein Osh4 in complex with 25-hydroxycholesterol | | Descriptor: | 25-HYDROXYCHOLESTEROL, KES1 protein | | Authors: | Im, Y.J, Raychaudhuri, S, Prinz, W.A, Hurley, J.H. | | Deposit date: | 2005-04-26 | | Release date: | 2005-09-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural mechanism for sterol sensing and transport by OSBP-related proteins
Nature, 437, 2005
|
|
1Z2N
 
 | | Inositol 1,3,4-trisphosphate 5/6-kinase complexed Mg2+/ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, inositol 1,3,4-trisphosphate 5/6-kinase | | Authors: | Miller, G.J, Wilson, M.P, Majerus, P.W, Hurley, J.H. | | Deposit date: | 2005-03-08 | | Release date: | 2005-04-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Specificity determinants in inositol polyphosphate synthesis: crystal structure of inositol 1,3,4-trisphosphate 5/6-kinase.
Mol.Cell, 18, 2005
|
|
1XHB
 
 | | The Crystal Structure of UDP-GalNAc: polypeptide alpha-N-acetylgalactosaminyltransferase-T1 | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, Polypeptide N-acetylgalactosaminyltransferase 1, ... | | Authors: | Fritz, T.A, Hurley, J.H, Trinh, L.B, Shiloach, J, Tabak, L.A. | | Deposit date: | 2004-09-17 | | Release date: | 2004-10-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The beginnings of mucin biosynthesis: The crystal structure of UDP-GalNAc:polypeptide {alpha}-N-acetylgalactosaminyltransferase-T1
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1Z2P
 
 | | Inositol 1,3,4-trisphosphate 5/6-Kinase in complex with Mg2+/AMP-PCP/Ins(1,3,4)P3 | | Descriptor: | (1S,3S,4S)-1,3,4-TRIPHOSPHO-MYO-INOSITOL, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Miller, G.J, Wilson, M.P, Majerus, P.W, Hurley, J.H. | | Deposit date: | 2005-03-08 | | Release date: | 2005-04-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Specificity determinants in inositol polyphosphate synthesis: crystal structure of inositol 1,3,4-trisphosphate 5/6-kinase.
Mol.Cell, 18, 2005
|
|
2OJQ
 
 | | Crystal structure of Alix V domain | | Descriptor: | Programmed cell death 6-interacting protein | | Authors: | Lee, S, Hurley, J.H. | | Deposit date: | 2007-01-13 | | Release date: | 2007-02-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Structural basis for viral late-domain binding to Alix
Nat.Struct.Mol.Biol., 14, 2007
|
|
1ZHW
 
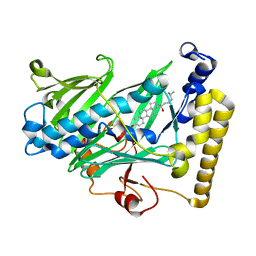 | | Structure of yeast oxysterol binding protein Osh4 in complex with 20-hydroxycholesterol | | Descriptor: | 20-HYDROXYCHOLESTEROL, KES1 protein, LEAD (II) ION | | Authors: | Im, Y.J, Raychaudhuri, S, Prinz, W.A, Hurley, J.H. | | Deposit date: | 2005-04-26 | | Release date: | 2005-09-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural mechanism for sterol sensing and transport by OSBP-related proteins
Nature, 437, 2005
|
|
1ZI7
 
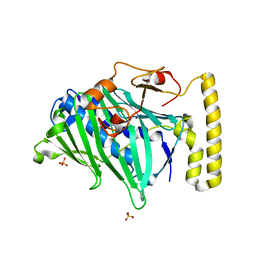 | | Structure of truncated yeast oxysterol binding protein Osh4 | | Descriptor: | KES1 protein, SULFATE ION | | Authors: | Im, Y.J, Raychaudhuri, S, Prinz, W.A, Hurley, J.H. | | Deposit date: | 2005-04-27 | | Release date: | 2005-09-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural mechanism for sterol sensing and transport by OSBP-related proteins
Nature, 437, 2005
|
|
1ZHZ
 
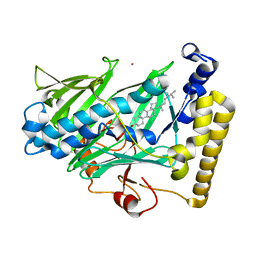 | | Structure of yeast oxysterol binding protein Osh4 in complex with ergosterol | | Descriptor: | ERGOSTEROL, KES1 protein, LEAD (II) ION | | Authors: | Im, Y.J, Raychaudhuri, S, Prinz, W.A, Hurley, J.H. | | Deposit date: | 2005-04-26 | | Release date: | 2005-09-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural mechanism for sterol sensing and transport by OSBP-related proteins
Nature, 437, 2005
|
|
1ZHY
 
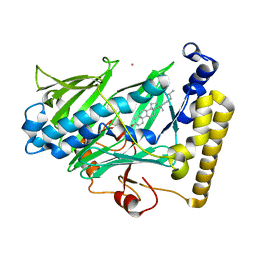 | | Structure of yeast oxysterol binding protein Osh4 in complex with cholesterol | | Descriptor: | CHOLESTEROL, KES1 protein, LEAD (II) ION | | Authors: | Im, Y.J, Raychaudhuri, S, Prinz, W.A, Hurley, J.H. | | Deposit date: | 2005-04-26 | | Release date: | 2005-09-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural mechanism for sterol sensing and transport by OSBP-related proteins
Nature, 437, 2005
|
|
1ZHT
 
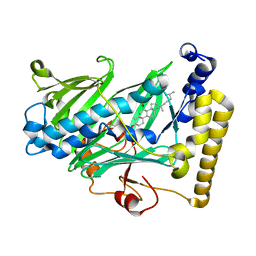 | | Structure of yeast oxysterol binding protein Osh4 in complex with 7-hydroxycholesterol | | Descriptor: | 7-HYDROXYCHOLESTEROL, KES1 protein | | Authors: | Im, Y.J, Raychaudhuri, S, Prinz, W.A, Hurley, J.H. | | Deposit date: | 2005-04-26 | | Release date: | 2005-09-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural mechanism for sterol sensing and transport by OSBP-related proteins
Nature, 437, 2005
|
|
1Z2O
 
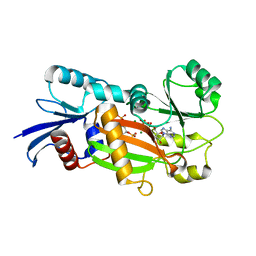 | | Inositol 1,3,4-trisphosphate 5/6-Kinase in complex with mg2+/ADP/Ins(1,3,4,6)P4 | | Descriptor: | (1S,3R,4R,6S)-1,3,4,6-TETRAPKISPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Miller, G.J, Wilson, M.P, Majerus, P.W, Hurley, J.H. | | Deposit date: | 2005-03-08 | | Release date: | 2005-04-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Specificity determinants in inositol polyphosphate synthesis: crystal structure of inositol 1,3,4-trisphosphate 5/6-kinase.
Mol.Cell, 18, 2005
|
|
1YD8
 
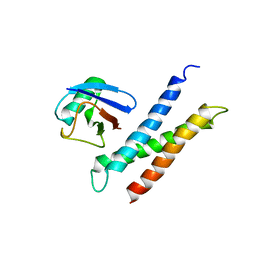 | | COMPLEX OF HUMAN GGA3 GAT DOMAIN AND UBIQUITIN | | Descriptor: | ADP-RIBOSYLATION FACTOR BINDING PROTEIN GGA3, UBIQUIN | | Authors: | Prag, G, Lee, S, Mattera, R, Arighi, C.N, Beach, B.M, Bonifacino, J.S, Hurley, J.H. | | Deposit date: | 2004-12-23 | | Release date: | 2005-02-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural mechanism for ubiquitinated-cargo recognition by the Golgi-localized, {gamma}-ear-containing, ADP-ribosylation-factor-binding proteins
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1ZB1
 
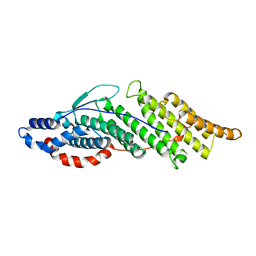 | | Structure basis for endosomal targeting by the Bro1 domain | | Descriptor: | BRO1 protein | | Authors: | Kim, J, Sitaraman, S, Hierro, A, Beach, B.M, Odorizzi, G, Hurley, J.H. | | Deposit date: | 2005-04-07 | | Release date: | 2005-06-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for endosomal targeting by the Bro1 domain.
Dev.Cell, 8, 2005
|
|
2P22
 
 | | Structure of the Yeast ESCRT-I Heterotetramer Core | | Descriptor: | Hypothetical 12.0 kDa protein in ADE3-SER2 intergenic region, Protein SRN2, SULFATE ION, ... | | Authors: | Kostelansky, M.S, Hurley, J.H. | | Deposit date: | 2007-03-06 | | Release date: | 2007-06-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular architecture and functional model of the complete yeast ESCRT-I heterotetramer.
Cell(Cambridge,Mass.), 129, 2007
|
|
1XA6
 
 | | Crystal Structure of the Human Beta2-Chimaerin | | Descriptor: | Beta2-chimaerin, ZINC ION | | Authors: | Canagarajah, B, Leskow, F.C, Ho, J.Y, Mischak, H, Saidi, L.F, Kazanietz, M.G, Hurley, J.H. | | Deposit date: | 2004-08-25 | | Release date: | 2004-11-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural mechanism for lipid activation of the Rac-specific GAP, beta2-chimaerin.
Cell(Cambridge,Mass.), 119, 2004
|
|
1I9Y
 
 | | CRYSTAL STRUCTURE OF INOSITOL POLYPHOSPHATE 5-PHOSPHATASE DOMAIN (IPP5C) OF SPSYNAPTOJANIN | | Descriptor: | PHOSPHATIDYLINOSITOL PHOSPHATE PHOSPHATASE | | Authors: | Tsujishita, Y, Guo, S, Stolz, L, York, J.D, Hurley, J.H. | | Deposit date: | 2001-03-21 | | Release date: | 2001-05-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Specificity determinants in phosphoinositide dephosphorylation: crystal structure of an archetypal inositol polyphosphate 5-phosphatase.
Cell(Cambridge,Mass.), 105, 2001
|
|
1P0Y
 
 | | Crystal structure of the SET domain of LSMT bound to MeLysine and AdoHcy | | Descriptor: | N-METHYL-LYSINE, Ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, chloroplast, ... | | Authors: | Trievel, R.C, Flynn, E.M, Houtz, R.L, Hurley, J.H. | | Deposit date: | 2003-04-11 | | Release date: | 2003-07-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Mechanism of multiple lysine methylation by the SET domain enzyme Rubisco LSMT
Nat.Struct.Biol., 10, 2003
|
|
